5QIK
 
 | | TGF-BETA RECEPTOR TYPE 1 KINASE DOMAIN (T204D) IN COMPLEX WITH N-{4-[3-(6-fluoropyridin-3-yl)-4-oxo-4,5,6,7-tetrahydro-1H-pyrrolo[3,2-c]pyridin-2-yl]pyridin-2-yl}acetamide | | Descriptor: | GLYCEROL, N-{4-[3-(6-fluoropyridin-3-yl)-4-oxo-4,5,6,7-tetrahydro-1H-pyrrolo[3,2-c]pyridin-2-yl]pyridin-2-yl}acetamide, TGF-beta receptor type-1 | | Authors: | Sheriff, S. | | Deposit date: | 2018-08-05 | | Release date: | 2018-10-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Discovery of 4-Azaindole Inhibitors of TGF beta RI as Immuno-oncology Agents.
ACS Med Chem Lett, 9, 2018
|
|
3P7J
 
 | | Drosophila HP1a chromo shadow domain | | Descriptor: | GLYCEROL, Heterochromatin protein 1 | | Authors: | Kim, D, Chruszcz, M, Minor, W, Khorasanizadeh, S. | | Deposit date: | 2010-10-12 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The HP1a Disordered C Terminus and Chromo Shadow Domain Cooperate to Select Target Peptide Partners.
Chembiochem, 12, 2011
|
|
3P4S
 
 | | Crystal structure of Menaquinol:fumarate oxidoreductase in complex with a 3-nitropropionate adduct | | Descriptor: | 3-NITROPROPANOIC ACID, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Tomasiak, T.M, Archuleta, T.L, Andrell, J, Luna-Chavez, C, Davis, T.A, Sarwar, M, Ham, A.J, McDonald, W.H, Yankowskaya, V, Stern, H.A, Johnston, J.N, Maklashina, E, Cecchini, G, Iverson, T.M. | | Deposit date: | 2010-10-07 | | Release date: | 2010-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Geometric restraint drives on- and off-pathway catalysis by the Escherichia coli menaquinol:fumarate reductase.
J.Biol.Chem., 286, 2011
|
|
4EPV
 
 | | Discovery of Small Molecules that Bind to K-Ras and Inhibit Sos-mediated Activation | | Descriptor: | 2-(1H-indol-3-ylmethyl)-1H-imidazo[4,5-c]pyridine, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Sun, Q, Burke, J.P, Phan, J, Burns, M.C, Olejniczak, E.T, Waterson, A.G, Lee, T, Rossanese, O.W, Fesik, S.W. | | Deposit date: | 2012-04-17 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Discovery of Small Molecules that Bind to K-Ras and Inhibit Sos-Mediated Activation.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
1TNP
 
 | |
2OC2
 
 | | Structure of testis ACE with RXPA380 | | Descriptor: | Angiotensin-converting enzyme, somatic isoform, CHLORIDE ION, ... | | Authors: | Corradi, H.R, Anthony, C.S, Schwager, S.L, Redelinghuys, P, Georgiadis, D, Dive, V, Acharya, K.R, Sturrock, E.D. | | Deposit date: | 2006-12-20 | | Release date: | 2007-05-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The structure of testis angiotensin-converting enzyme in complex with the C domain-specific inhibitor RXPA380.
Biochemistry, 46, 2007
|
|
4OK3
 
 | |
4OK6
 
 | |
1TEC
 
 | | CRYSTALLOGRAPHIC REFINEMENT BY INCORPORATION OF MOLECULAR DYNAMICS. THE THERMOSTABLE SERINE PROTEASE THERMITASE COMPLEXED WITH EGLIN-C | | Descriptor: | CALCIUM ION, EGLIN C, SODIUM ION, ... | | Authors: | Gros, P, Dijkstra, B.W, Hol, W.G.J. | | Deposit date: | 1989-05-24 | | Release date: | 1989-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic refinement by incorporation of molecular dynamics: thermostable serine protease thermitase complexed with eglin c.
Acta Crystallogr.,Sect.B, 45, 1989
|
|
4F4X
 
 | | Y-family DNA polymerase chimera Dbh-Dpo4-Dpo4 #2 | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*GP*GP*CP*AP*CP*TP*GP*AP*TP*CP*GP*GP*GP*C)-3'), ... | | Authors: | Pata, J.D, Wilson, R.C. | | Deposit date: | 2012-05-11 | | Release date: | 2013-01-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.049 Å) | | Cite: | Y-family polymerase conformation is a major determinant of fidelity and translesion specificity.
Structure, 21, 2013
|
|
5OGI
 
 | | Complex of a binding protein and human adenovirus C 5 hexon | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Schmid, M, Ernst, P, Honegger, A, Suomalainen, M, Zimmermann, M, Braun, L, Stauffer, S, Thom, C, Dreier, B, Eibauer, M, Kipar, A, Vogel, V, Greber, U.F, Medalia, O, Plueckthun, A. | | Deposit date: | 2017-07-13 | | Release date: | 2018-02-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Adenoviral vector with shield and adapter increases tumor specificity and escapes liver and immune control.
Nat Commun, 9, 2018
|
|
4F4W
 
 | | Y-family DNA polymerase chimera Dbh-Dpo4-Dpo4 #1 | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*GP*GP*CP*AP*CP*TP*GP*AP*TP*CP*GP*GP*C)-3'), ... | | Authors: | Pata, J.D, Wilson, R.C. | | Deposit date: | 2012-05-11 | | Release date: | 2013-01-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Y-family polymerase conformation is a major determinant of fidelity and translesion specificity.
Structure, 21, 2013
|
|
1PIC
 
 | | PHOSPHATIDYLINOSITOL 3-KINASE, P85-ALPHA SUBUNIT: C-TERMINAL SH2 DOMAIN COMPLEXED WITH A TYR751 PHOSPHOPEPTIDE FROM THE PDGF RECEPTOR, NMR, MINIMIZED MEAN STRUCTURE | | Descriptor: | BETA-PLATELET-DERIVED GROWTH FACTOR RECEPTOR, PHOSPHATIDYLINOSITOL 3-KINASE | | Authors: | Breeze, A.L, Kara, B.V, Barratt, D.G, Anderson, M, Smith, J.C, Luke, R.W, Best, J.R, Cartlidge, S.A. | | Deposit date: | 1997-06-23 | | Release date: | 1997-09-17 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structure of a specific peptide complex of the carboxy-terminal SH2 domain from the p85 alpha subunit of phosphatidylinositol 3-kinase.
EMBO J., 15, 1996
|
|
1CTF
 
 | |
1CMQ
 
 | | SMALL MOLECULE BINDING TO AN ARTIFICIALLY CREATED CAVITY AT THE ACTIVE SITE OF CYTOCHROME C PEROXIDASE | | Descriptor: | CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Fitzgerald, M.M, Mcree, D.E, Churchill, M.J, Goodin, D.B. | | Deposit date: | 1993-11-23 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Small molecule binding to an artificially created cavity at the active site of cytochrome c peroxidase.
Biochemistry, 33, 1994
|
|
3SGD
 
 | | Crystal structure of the mouse mAb 17.2 | | Descriptor: | CALCIUM ION, Heavy Chain, Light Chain | | Authors: | Pizarro, J.C, Boulot, G, Hontebeyrie, M, Bentley, G.A. | | Deposit date: | 2011-06-14 | | Release date: | 2011-11-09 | | Last modified: | 2013-01-23 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal structure of the complex mAb 17.2 and the C-terminal region of Trypanosoma cruzi P2 Beta protein: implications in cross-reactivity
Plos Negl Trop Dis, 5, 2011
|
|
1UKK
 
 | | Structure of Osmotically Inducible Protein C from Thermus thermophilus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Osmotically Inducible Protein C | | Authors: | Rehse, P.H, Kuramitsu, S, Yokoyama, S, Miyano, M, Tahirov, T.H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-08-24 | | Release date: | 2004-05-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic Structure and Biochemical Analysis of the Thermus thermophilus Osmotically Inducible Protein C
J.MOL.BIOL., 338, 2004
|
|
2PLE
 
 | | NUCLEAR MAGNETIC RESONANCE STRUCTURE OF AN SH2 DOMAIN OF PHOSPHOLIPASE C-GAMMA1 COMPLEXED WITH A HIGH AFFINITY BINDING PEPTIDE | | Descriptor: | PHOSPHOLIPASE C GAMMA-1, C-TERMINAL SH2 DOMAIN, PHOSPHOPEPTIDE FROM PDGF | | Authors: | Pascal, S.M, Singer, A.U, Gish, G, Yamazaki, T, Shoelson, S.E, Pawson, T, Kay, L.E, Forman-Kay, J.D. | | Deposit date: | 1994-08-19 | | Release date: | 1995-01-26 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure of an SH2 domain of phospholipase C-gamma 1 complexed with a high affinity binding peptide.
Cell(Cambridge,Mass.), 77, 1994
|
|
1CMP
 
 | | SMALL MOLECULE BINDING TO AN ARTIFICIALLY CREATED CAVITY AT THE ACTIVE SITE OF CYTOCHROME C PEROXIDASE | | Descriptor: | 2,3-DIMETHYLIMIDAZOLIUM ION, CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Fitzgerald, M.M, Mcree, D.E, Churchill, M.J, Goodin, D.B. | | Deposit date: | 1993-11-23 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Small molecule binding to an artificially created cavity at the active site of cytochrome c peroxidase.
Biochemistry, 33, 1994
|
|
2Q5Y
 
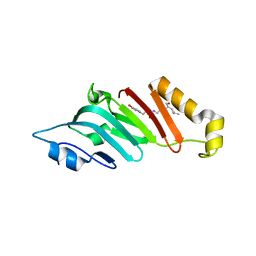 | | Crystal Structure of the C-terminal domain of hNup98 | | Descriptor: | Nuclear pore complex protein Nup96, Nuclear pore complex protein Nup98 | | Authors: | Sun, Y, Guo, H.C. | | Deposit date: | 2007-06-03 | | Release date: | 2008-10-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural constraints on autoprocessing of the human nucleoporin Nup98.
Protein Sci., 17, 2008
|
|
2BNW
 
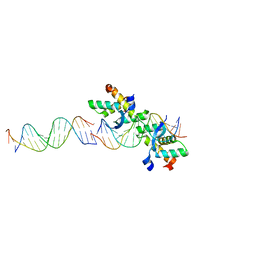 | | Structural basis for cooperative binding of Ribbon-Helix-Helix Omega repressor to direct DNA heptad repeats | | Descriptor: | 5'-D(*CP*TP*TP*GP*TP*GP*AP*TP*TP*TP *GP*TP*GP*AP*TP*TP*CP*G)-3', 5'-D(*GP*AP*AP*TP*CP*AP*CP*AP*AP*AP *TP*CP*AP*CP*AP*AP*GP*C)-3', ORF OMEGA | | Authors: | Weihofen, W.A, Cicek, A, Pratto, F, Alonso, J.C, Saenger, W. | | Deposit date: | 2005-04-05 | | Release date: | 2006-03-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structures of Omega Repressors Bound to Direct and Inverted DNA Repeats Explain Modulation of Transcription.
Nucleic Acids Res., 34, 2006
|
|
4K08
 
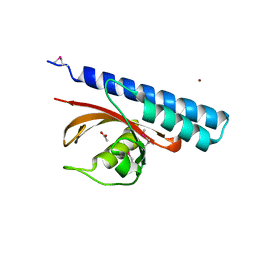 | | Periplasmic sensor domain of chemotaxis protein, Adeh_3718 | | Descriptor: | ACETATE ION, Methyl-accepting chemotaxis sensory transducer, ZINC ION | | Authors: | Pokkuluri, P.R, Mack, J.C, Bearden, J, Rakowski, E, Schiffer, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-04-03 | | Release date: | 2013-07-17 | | Last modified: | 2015-04-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Analysis of periplasmic sensor domains from Anaeromyxobacter dehalogenans 2CP-C: structure of one sensor domain from a histidine kinase and another from a chemotaxis protein.
Microbiologyopen, 2, 2013
|
|
3PA8
 
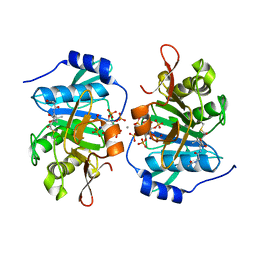 | |
1A2I
 
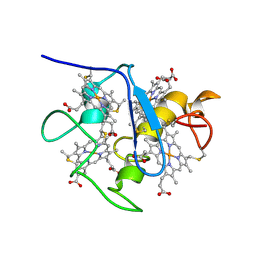 | | SOLUTION STRUCTURE OF DESULFOVIBRIO VULGARIS (HILDENBOROUGH) FERROCYTOCHROME C3, NMR, 20 STRUCTURES | | Descriptor: | CYTOCHROME C3, HEME C | | Authors: | Messias, A.C, Kastrau, D.H.K, Costa, H.S, Legall, J, Turner, D.L, Santos, H, Xavier, A.V. | | Deposit date: | 1998-01-05 | | Release date: | 1998-07-08 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Desulfovibrio vulgaris (Hildenborough) ferrocytochrome c3: structural basis for functional cooperativity.
J.Mol.Biol., 281, 1998
|
|
2PLD
 
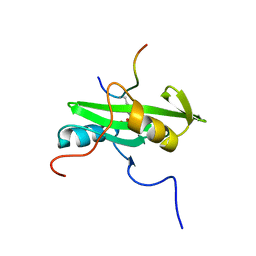 | | NUCLEAR MAGNETIC RESONANCE STRUCTURE OF AN SH2 DOMAIN OF PHOSPHOLIPASE C-GAMMA1 COMPLEXED WITH A HIGH AFFINITY BINDING PEPTIDE | | Descriptor: | PHOSPHOLIPASE C GAMMA-1, C-TERMINAL SH2 DOMAIN, PHOSPHOPEPTIDE FROM PDGF | | Authors: | Pascal, S.M, Singer, A.U, Gish, G, Yamazaki, T, Shoelson, S.E, Pawson, T, Kay, L.E, Forman-Kay, J.D. | | Deposit date: | 1994-08-19 | | Release date: | 1995-01-26 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure of an SH2 domain of phospholipase C-gamma 1 complexed with a high affinity binding peptide.
Cell(Cambridge,Mass.), 77, 1994
|
|
