5FJ9
 
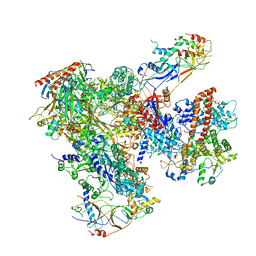 | | Cryo-EM structure of yeast apo RNA polymerase III at 4.6 A | | Descriptor: | DNA-DIRECTED RNA POLYMERASE III SUBUNIT RPC1, DNA-DIRECTED RNA POLYMERASE III SUBUNIT RPC10, DNA-DIRECTED RNA POLYMERASE III SUBUNIT RPC2, ... | | Authors: | Hoffmann, N.A, Jakobi, A.J, Moreno-Morcillo, M, Glatt, S, Kosinski, J, Hagen, W.J, Sachse, C, Muller, C.W. | | Deposit date: | 2015-10-06 | | Release date: | 2015-11-25 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Molecular Structures of Unbound and Transcribing RNA Polymerase III.
Nature, 528, 2015
|
|
5FJA
 
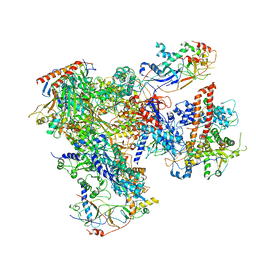 | | Cryo-EM structure of yeast RNA polymerase III at 4.7 A | | Descriptor: | DNA-DIRECTED RNA POLYMERASE III SUBUNIT RPC1, DNA-DIRECTED RNA POLYMERASE III SUBUNIT RPC10, DNA-DIRECTED RNA POLYMERASE III SUBUNIT RPC2, ... | | Authors: | Hoffmann, N.A, Jakobi, A.J, Moreno-Morcillo, M, Glatt, S, Kosinski, J, Hagen, W.J, Sachse, C, Muller, C.W. | | Deposit date: | 2015-10-06 | | Release date: | 2015-11-25 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.65 Å) | | Cite: | Molecular Structures of Unbound and Transcribing RNA Polymerase III.
Nature, 528, 2015
|
|
6RFL
 
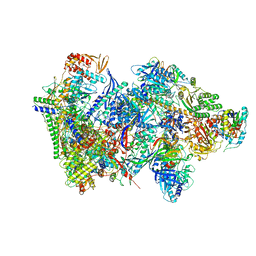 | | Structure of the complete Vaccinia DNA-dependent RNA polymerase complex | | Descriptor: | DNA-dependent RNA polymerase subunit rpo132, DNA-dependent RNA polymerase subunit rpo147, DNA-dependent RNA polymerase subunit rpo18, ... | | Authors: | Grimm, C, Hillen, S.H, Bedenk, K, Bartuli, J, Neyer, S, Zhang, Q, Huettenhofer, A, Erlacher, M, Dienemann, C, Schlosser, A, Urlaub, H, Boettcher, B, Szalay, A.A, Cramer, P, Fischer, U. | | Deposit date: | 2019-04-15 | | Release date: | 2019-12-11 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structural Basis of Poxvirus Transcription: Vaccinia RNA Polymerase Complexes.
Cell, 179, 2019
|
|
7VBA
 
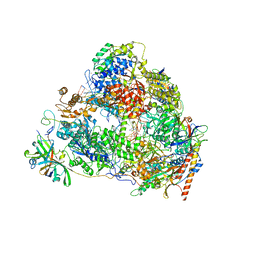 | | Structure of the pre state human RNA Polymerase I Elongation Complex | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]cytidine, DNA (5'-D(P*A*CP*TP*GP*TP*CP*CP*TP*CP*TP*GP*GP*C)-3'), DNA (5'-D(P*GP*CP*CP*AP*GP*AP*GP*AP*CP*AP*GP*CP*GP*AP*GP*TP*CP*AP*GP*CP*AP*A)-3'), ... | | Authors: | Zhao, D, Liu, W, Chen, K, Yang, H, Xu, Y. | | Deposit date: | 2021-08-31 | | Release date: | 2022-02-16 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structure of the human RNA polymerase I elongation complex.
Cell Discov, 7, 2021
|
|
7VBC
 
 | | Back track state of human RNA Polymerase I Elongation Complex | | Descriptor: | DNA (5'-D(*GP*TP*AP*CP*TP*GP*TP*CP*CP*TP*CP*TP*GP*G)-3'), DNA (5'-D(P*AP*GP*GP*AP*CP*AP*GP*CP*GP*TP*GP*TP*CP*AP*GP*CP*AP*AP*TP*A)-3'), DNA-directed RNA polymerase I subunit RPA1, ... | | Authors: | Zhao, D, Liu, W, Chen, K, Yang, H, Xu, Y. | | Deposit date: | 2021-08-31 | | Release date: | 2022-02-16 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structure of the human RNA polymerase I elongation complex.
Cell Discov, 7, 2021
|
|
7VBB
 
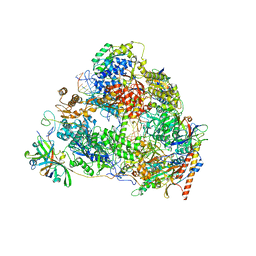 | | Structure of the post state human RNA Polymerase I Elongation Complex | | Descriptor: | DNA (25-MER), DNA (5'-D(*CP*TP*GP*TP*CP*CP*TP*CP*TP*GP*GP*CP*GP*A)-3'), DNA-directed RNA polymerase I subunit RPA1, ... | | Authors: | Zhao, D, Liu, W, Chen, K, Yang, H, Xu, Y. | | Deposit date: | 2021-08-31 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Structure of the human RNA polymerase I elongation complex.
Cell Discov, 7, 2021
|
|
8C8H
 
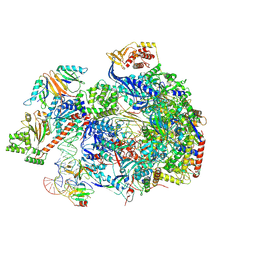 | |
7DTH
 
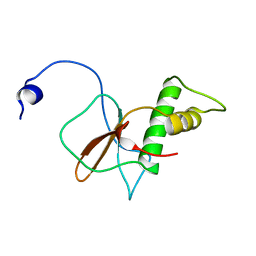 | | Solution structure of RPB6, common subunit of RNA polymerases I, II, and III | | Descriptor: | DNA-directed RNA polymerases I, II, and III subunit RPABC2 | | Authors: | Okuda, M, Nishimura, Y. | | Deposit date: | 2021-01-05 | | Release date: | 2022-01-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Three human RNA polymerases interact with TFIIH via a common RPB6 subunit.
Nucleic Acids Res., 50, 2022
|
|
7DTI
 
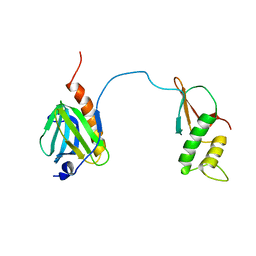 | |
8A43
 
 | | Human RNA polymerase I | | Descriptor: | DNA-directed RNA polymerase I subunit RPA1, DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA2, ... | | Authors: | Daiss, J.L, Pilsl, M, Straub, K, Bleckmann, A, Hoecherl, M, Heiss, F.B, Abascal-Palacios, G, Ramsay, E, Tluckova, K, Mars, J.C, Fuertges, T, Bruckmann, A, Rudack, T, Bernecky, C, Lamour, V, Panov, K, Vannini, A, Moss, T, Engel, C. | | Deposit date: | 2022-06-10 | | Release date: | 2022-08-31 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.09 Å) | | Cite: | The human RNA polymerase I structure reveals an HMG-like docking domain specific to metazoans.
Life Sci Alliance, 5, 2022
|
|
7RIL
 
 | | Crystal structure of hairpin polyamide Py-Im 1 bound to 5' CCTGACCAGG | | Descriptor: | 3-({3-[(3-{[4-({4-[(4-{[4-({(2R)-2-amino-4-[(1-methyl-4-{[1-methyl-4-({1-methyl-4-[(1-methyl-1H-imidazole-2-carbonyl)amino]-1H-imidazole-2-carbonyl}amino)-1H-pyrrole-2-carbonyl]amino}-1H-pyrrole-2-carbonyl)amino]butanoyl}amino)-1-methyl-1H-imidazole-2-carbonyl]amino}-1-methyl-1H-pyrrole-2-carbonyl)amino]-1-methyl-1H-pyrrole-2-carbonyl}amino)-1-methyl-1H-pyrrole-2-carbonyl]amino}propyl)(methyl)amino]propyl}carbamoyl)benzoic acid, ACETATE ION, non-template DNA, ... | | Authors: | Oh, J, Dervan, P.B, Wang, D. | | Deposit date: | 2021-07-20 | | Release date: | 2022-01-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | RNA polymerase II trapped on a molecular treadmill: Structural basis of persistent transcriptional arrest by a minor groove DNA binder.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8P2I
 
 | | Cryo-EM structure of Pyrococcus furiosus apo form RNA polymerase contracted clamp conformation with Spt4/5 | | Descriptor: | DNA-directed RNA polymerase subunit Rpo10, DNA-directed RNA polymerase subunit Rpo11, DNA-directed RNA polymerase subunit Rpo12, ... | | Authors: | Tarau, D.M, Reichelt, R, Heiss, F.B, Pilsl, M, Hausner, W, Engel, C, Grohmann, D. | | Deposit date: | 2023-05-16 | | Release date: | 2024-04-24 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of archaeal RNA polymerase transcription elongation and Spt4/5 recruitment.
Nucleic Acids Res., 52, 2024
|
|
6KF9
 
 | | Cryo-EM structure of Thermococcus kodakarensis RNA polymerase | | Descriptor: | DNA (27-MER), DNA (5'-D(P*TP*CP*GP*GP*TP*AP*AP*TP*CP*AP*CP*GP*CP*TP*CP*C)-3'), DNA-directed RNA polymerase subunit, ... | | Authors: | Jun, S.-H, Hyun, J, Jeong, H, Cha, J.S, Kim, H, Bartlett, M.S, Cho, H.-S, Murakami, K.S. | | Deposit date: | 2019-07-07 | | Release date: | 2020-07-01 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Direct binding of TFE alpha opens DNA binding cleft of RNA polymerase.
Nat Commun, 11, 2020
|
|
6KF4
 
 | | Cryo-EM structure of Thermococcus kodakarensis RNA polymerase | | Descriptor: | DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit A'', DNA-directed RNA polymerase subunit D, ... | | Authors: | Jun, S.-H, Hyun, J, Jeong, H, Cha, J.S, Kim, H, Bartlett, M.S, Cho, H.-S, Murakami, K.S. | | Deposit date: | 2019-07-06 | | Release date: | 2020-07-01 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.97 Å) | | Cite: | Direct binding of TFE alpha opens DNA binding cleft of RNA polymerase.
Nat Commun, 11, 2020
|
|
8Q3B
 
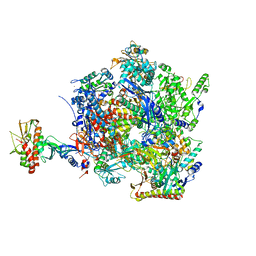 | | The closed state of the ASFV apo-RNA polymerase | | Descriptor: | DNA-directed RNA polymerase RPB1 homolog, DNA-directed RNA polymerase RPB10 homolog, DNA-directed RNA polymerase RPB2 homolog, ... | | Authors: | Pilotto, S, Sykora, M, Cackett, G, Werner, F. | | Deposit date: | 2023-08-03 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Structure of the recombinant RNA polymerase from African Swine Fever Virus.
Nat Commun, 15, 2024
|
|
8Q3K
 
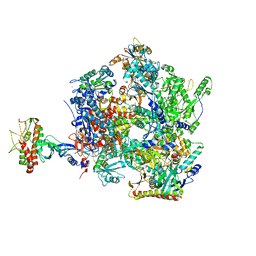 | | The open state of the ASFV apo-RNA polymerase | | Descriptor: | DNA-directed RNA polymerase RPB1 homolog, DNA-directed RNA polymerase RPB10 homolog, DNA-directed RNA polymerase RPB2 homolog, ... | | Authors: | Pilotto, S, Sykora, M, Cackett, G, Werner, F. | | Deposit date: | 2023-08-04 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Structure of the recombinant RNA polymerase from African Swine Fever Virus.
Nat Commun, 15, 2024
|
|
8ORQ
 
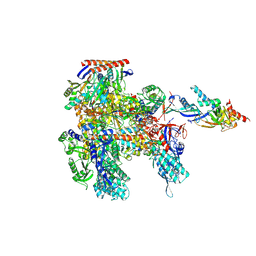 | | Cryo-EM structure of Pyrococcus furiosus apo form RNA polymerase open clamp conformation | | Descriptor: | DNA-directed RNA polymerase subunit Rpo10, DNA-directed RNA polymerase subunit Rpo11, DNA-directed RNA polymerase subunit Rpo12, ... | | Authors: | Tarau, D.M, Reichelt, R, Heiss, F.B, Pilsl, M, Hausner, W, Engel, C, Grohmann, D. | | Deposit date: | 2023-04-17 | | Release date: | 2024-04-24 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of archaeal RNA polymerase transcription elongation and Spt4/5 recruitment.
Nucleic Acids Res., 52, 2024
|
|
7AST
 
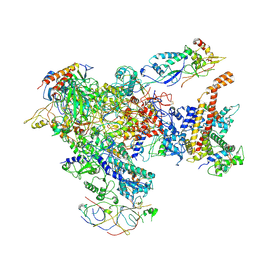 | | Apo Human RNA Polymerase III | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Ramsay, E.P, Abascal-Palacios, G, Daiss, J.L, King, H, Gouge, J, Pilsl, M, Beuron, F, Morris, E, Gunkel, P, Engel, C, Vannini, A. | | Deposit date: | 2020-10-28 | | Release date: | 2020-12-23 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of human RNA polymerase III.
Nat Commun, 11, 2020
|
|
1ARO
 
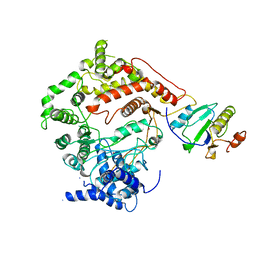 | | T7 RNA POLYMERASE COMPLEXED WITH T7 LYSOZYME | | Descriptor: | MERCURY (II) ION, T7 LYSOZYME, T7 RNA POLYMERASE | | Authors: | Steitz, T, Jeruzalmi, D. | | Deposit date: | 1997-08-08 | | Release date: | 1998-10-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of T7 RNA polymerase complexed to the transcriptional inhibitor T7 lysozyme.
EMBO J., 17, 1998
|
|
7MKQ
 
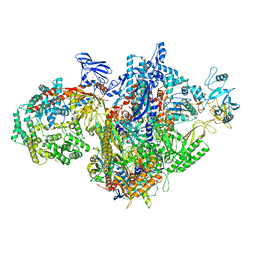 | | Escherichia coli RNA polymerase and RapA binary complex | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Qayyum, M.Z, Murakami, K.S. | | Deposit date: | 2021-04-26 | | Release date: | 2021-06-02 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural basis of RNA polymerase recycling by the Swi2/Snf2 family of ATPase RapA in Escherichia coli.
J.Biol.Chem., 297, 2021
|
|
7MKN
 
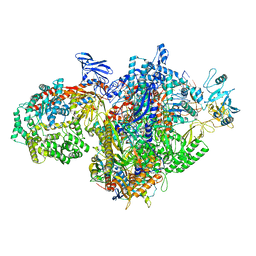 | | Escherichia coli RNA polymerase and RapA elongation complex | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]cytidine, DNA (29-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Qayyum, M.Z, Murakami, K.S. | | Deposit date: | 2021-04-26 | | Release date: | 2021-06-02 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of RNA polymerase recycling by the Swi2/Snf2 family of ATPase RapA in Escherichia coli.
J.Biol.Chem., 297, 2021
|
|
2PMZ
 
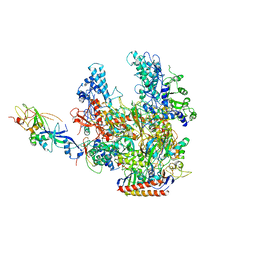 | | Archaeal RNA polymerase from Sulfolobus solfataricus | | Descriptor: | DNA-directed RNA polymerase subunit A, DNA-directed RNA polymerase subunit A", DNA-directed RNA polymerase subunit B, ... | | Authors: | Murakami, K.S. | | Deposit date: | 2007-04-23 | | Release date: | 2008-02-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The X-ray crystal structure of RNA polymerase from Archaea
Nature, 451, 2008
|
|
1UVJ
 
 | | The structural basis for RNA specificity and Ca2 inhibition of an RNA-dependent RNA polymerase phi6p2 with 7nt RNA | | Descriptor: | 5'-R(*UP*UP*CP*CP)-3', MANGANESE (II) ION, RNA-directed RNA polymerase | | Authors: | Salgado, P.S, Makeyev, E.V, Butcher, S, Bamford, D, Stuart, D.I, Grimes, J.M. | | Deposit date: | 2004-01-21 | | Release date: | 2004-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural basis for RNA specificity and Ca2+ inhibition of an RNA-dependent RNA polymerase.
Structure, 12, 2004
|
|
1UVI
 
 | | The structural basis for RNA specificity and Ca2 inhibition of an RNA-dependent RNA polymerase phi6p2 with 6nt RNA | | Descriptor: | 5'-R(*UP*UP*UP*UP*CP*CP)-3', MANGANESE (II) ION, RNA-directed RNA polymerase | | Authors: | Salgado, P.S, Makeyev, E.V, Butcher, S, Bamford, D, Stuart, D.I, Grimes, J.M. | | Deposit date: | 2004-01-21 | | Release date: | 2004-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The structural basis for RNA specificity and Ca2+ inhibition of an RNA-dependent RNA polymerase.
Structure, 12, 2004
|
|
1UVN
 
 | | The structural basis for RNA specificity and Ca2 inhibition of an RNA-dependent RNA polymerase phi6p2 ca2+ inhibition complex | | Descriptor: | 5'-R(*UP*UP*UP*UP*CP*CP)-3', CALCIUM ION, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Salgado, P.S, Makeyev, E.V, Butcher, S, Bamford, D, Stuart, D.I, Grimes, J.M. | | Deposit date: | 2004-01-21 | | Release date: | 2004-02-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structural basis for RNA specificity and Ca2+ inhibition of an RNA-dependent RNA polymerase.
Structure, 12, 2004
|
|
