9PDD
 
 | | 22bin20S complex (NSF-alphaSNAP-2:2 syntaxin-1a:SNAP-25), hydrolyzing, class 29 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Alpha-soluble NSF attachment protein, ... | | Authors: | White, K.I, Brunger, A.T. | | Deposit date: | 2025-06-30 | | Release date: | 2025-08-06 | | Last modified: | 2025-10-08 | | Method: | ELECTRON MICROSCOPY (4.16 Å) | | Cite: | Structural remodeling of target-SNARE protein complexes by NSF enables synaptic transmission.
Nat Commun, 16, 2025
|
|
7Q06
 
 | | Crystal structure of TPADO in complex with 2-OH-TPA | | Descriptor: | 2-Hydroxyterephthalic acid, FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Zahn, M, Kincannon, W.M, DuBois, J.L, McGeehan, J.E. | | Deposit date: | 2021-10-14 | | Release date: | 2022-03-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Biochemical and structural characterization of an aromatic ring-hydroxylating dioxygenase for terephthalic acid catabolism.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Q05
 
 | | Crystal structure of TPADO in complex with TPA | | Descriptor: | FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, Lysozyme, ... | | Authors: | Zahn, M, Kincannon, W.M, DuBois, J.L, McGeehan, J.E. | | Deposit date: | 2021-10-14 | | Release date: | 2022-03-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Biochemical and structural characterization of an aromatic ring-hydroxylating dioxygenase for terephthalic acid catabolism.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Q04
 
 | | Crystal structure of TPADO in a substrate-free state | | Descriptor: | FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, Lysozyme, ... | | Authors: | Zahn, M, Kincannon, W.M, DuBois, J.L, McGeehan, J.E. | | Deposit date: | 2021-10-14 | | Release date: | 2022-04-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.281 Å) | | Cite: | Biochemical and structural characterization of an aromatic ring-hydroxylating dioxygenase for terephthalic acid catabolism.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
3PXP
 
 | |
8QWJ
 
 | |
4EVI
 
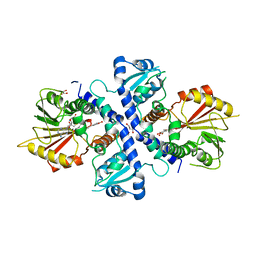 | | Crystal Structure Analysis of Coniferyl Alcohol 9-O-Methyltransferase from Linum Nodiflorum in Complex with Coniferyl Alcohol 9-Methyl Ether and S -Adenosyl-L-Homocysteine | | Descriptor: | 2-methoxy-4-[(1E)-3-methoxyprop-1-en-1-yl]phenol, 4-[(1E)-3-hydroxyprop-1-en-1-yl]-2-methoxyphenol, Coniferyl alcohol 9-O-methyltransferase, ... | | Authors: | Wolters, S, Heine, A, Petersen, M. | | Deposit date: | 2012-04-26 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.015 Å) | | Cite: | Structural analysis of coniferyl alcohol 9-O-methyltransferase from Linum nodiflorum reveals a novel active-site environment.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6MV1
 
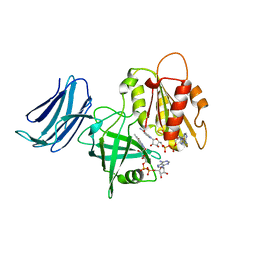 | | 2.15A resolution structure of the CS-b5R domains of human Ncb5or (NAD+ form) | | Descriptor: | Cytochrome b5 reductase 4, FLAVIN-ADENINE DINUCLEOTIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Benson, D.R, Cooper, A, Gao, P, Zhu, H. | | Deposit date: | 2018-10-24 | | Release date: | 2019-07-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structures of the naturally fused CS and cytochrome b5reductase (b5R) domains of Ncb5or reveal an expanded CS fold, extensive CS-b5R interactions and productive binding of the NAD(P)+nicotinamide ring.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
8QHN
 
 | |
6MV2
 
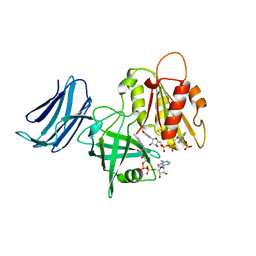 | | 2.05A resolution structure of the CS-b5R domains of human Ncb5or (NADP+ form) | | Descriptor: | Cytochrome b5 reductase 4, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Benson, D.R, Cooper, A, Gao, P, Zhu, H. | | Deposit date: | 2018-10-24 | | Release date: | 2019-07-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of the naturally fused CS and cytochrome b5reductase (b5R) domains of Ncb5or reveal an expanded CS fold, extensive CS-b5R interactions and productive binding of the NAD(P)+nicotinamide ring.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
5YGS
 
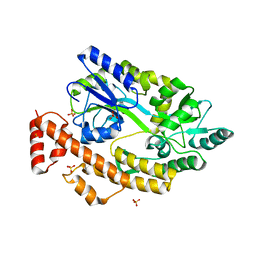 | | Human TNFRSF25 death domain | | Descriptor: | Human TNRSF25 death domain, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Yin, X, Jin, T. | | Deposit date: | 2017-09-26 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.691 Å) | | Cite: | Crystal structure and activation mechanism of DR3 death domain.
Febs J., 286, 2019
|
|
6KFB
 
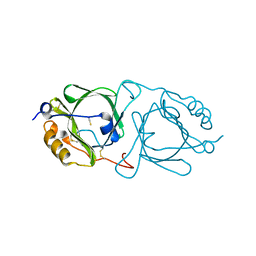 | | Hydroxynitrile lyase from the millipede, Chamberlinius hualienensis bound with thiocyanate | | Descriptor: | Hydroxynitrile lyase, THIOCYANATE ION, alpha-L-fucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Motojima, F, Izumi, A, Asano, Y. | | Deposit date: | 2019-07-07 | | Release date: | 2020-07-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | R-hydroxynitrile lyase from the cyanogenic millipede, Chamberlinius hualienensis-A new entry to the carrier protein family Lipocalines.
Febs J., 288, 2021
|
|
2R6S
 
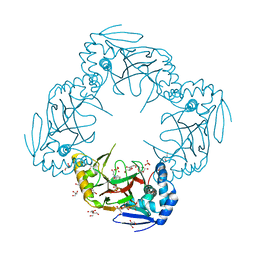 | | Crystal structure of Gab protein | | Descriptor: | BICINE, FE (II) ION, GLYCEROL, ... | | Authors: | Lohkamp, B, Dobritzsch, D. | | Deposit date: | 2007-09-06 | | Release date: | 2008-06-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A mixture of fortunes: the curious determination of the structure of Escherichia coli BL21 Gab protein.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
5Z5O
 
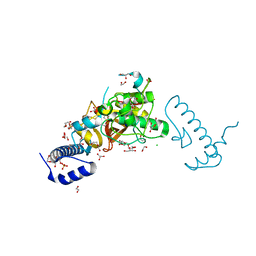 | | Structure of Pycnonodysostosis disease related I249T mutant of human cathepsin K | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Biswas, S, Roy, S. | | Deposit date: | 2018-01-19 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Not all pycnodysostosis-related mutants of human cathepsin K are inactive - crystal structure and biochemical studies of an active mutant I249T.
FEBS J., 285, 2018
|
|
3SOI
 
 | | Crystallographic structure of Bacillus licheniformis beta-lactamase W210F/W229F/W251F at 1.73 angstrom resolution | | Descriptor: | Beta-lactamase, CITRIC ACID | | Authors: | Acierno, J.P, Capaldi, S, Risso, V.A, Monaco, H.L, Ermacora, M.R. | | Deposit date: | 2011-06-30 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.729 Å) | | Cite: | X-ray evidence of a native state with increased compactness populated by tryptophan-less B. licheniformis beta-lactamase.
Protein Sci., 21, 2012
|
|
6NEE
 
 | |
8TXS
 
 | |
6NKQ
 
 | |
5GVY
 
 | | Crystal structure of SALT protein from Oryza sativa | | Descriptor: | Salt stress-induced protein, alpha-D-mannopyranose | | Authors: | Sharma, P, Sagar, A, Kaur, N, Sharma, I, Kirat, K, Ashish, F.N.U, Pati, P.K. | | Deposit date: | 2016-09-07 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.662 Å) | | Cite: | Structural insights into rice SalTol QTL located SALT protein.
Sci Rep, 10, 2020
|
|
5ZQV
 
 | |
3KW0
 
 | |
6XR3
 
 | |
3N2X
 
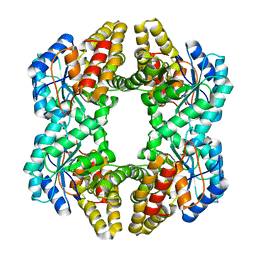 | | Crystal structure of YagE, a prophage protein belonging to the dihydrodipicolinic acid synthase family from E. coli K12 in complex with pyruvate | | Descriptor: | 1,2-ETHANEDIOL, Uncharacterized protein yagE | | Authors: | Bhaskar, V, Kumar, P.M, Manicka, S, Krishnaswamy, S. | | Deposit date: | 2010-05-19 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of biochemical and putative biological role of a xenolog from Escherichia coli using structural analysis.
Proteins, 79, 2011
|
|
9KZ1
 
 | | Crystal structure of PlySb from a chimeolysin ClyR | | Descriptor: | N-acetylmuramoyl-L-alanine amidase | | Authors: | Hu, F. | | Deposit date: | 2024-12-09 | | Release date: | 2025-10-22 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Domain Interactions in a Chimeric Dual-domain Lysin Lead to Broad Bactericidal Activity.
J.Mol.Biol., 437, 2025
|
|
3NEV
 
 | | Crystal structure of YagE, a prophage protein from E. coli K12 in complex with KDGal | | Descriptor: | 1,2-ETHANEDIOL, 3-DEOXY-D-LYXO-HEXONIC ACID, Uncharacterized protein yagE | | Authors: | Bhaskar, V, Kumar, P.M, Manicka, S, Krishnaswamy, S. | | Deposit date: | 2010-06-09 | | Release date: | 2011-04-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Identification of biochemical and putative biological role of a xenolog from Escherichia coli using structural analysis.
Proteins, 79, 2011
|
|
