5FXX
 
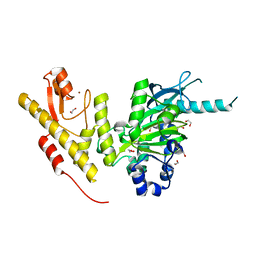 | | Crystal structure of JmjC domain of human histone demethylase UTY in complex with oxaloacetate | | Descriptor: | 1,2-ETHANEDIOL, HISTONE DEMETHYLASE UTY, MANGANESE (II) ION, ... | | Authors: | Nowak, R, Krojer, T, Johansson, C, Gileadi, C, Kupinska, K, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U. | | Deposit date: | 2016-03-03 | | Release date: | 2016-03-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal Structure of Jmjc Domain of Human Histone Demethylase Uty in Complex with Oxaloacetate
To be Published
|
|
8CLQ
 
 | |
8V78
 
 | |
8V75
 
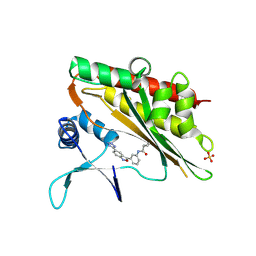 | |
8RHS
 
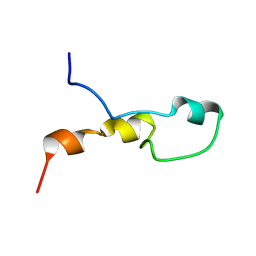 | |
8TL9
 
 | | Human Type 3 IP3 Receptor - Resting State (+IP3/ATP) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, ... | | Authors: | Paknejad, N, Sapuru, V, Hite, R.K. | | Deposit date: | 2023-07-26 | | Release date: | 2023-11-08 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural titration reveals Ca 2+ -dependent conformational landscape of the IP 3 receptor.
Nat Commun, 14, 2023
|
|
8V73
 
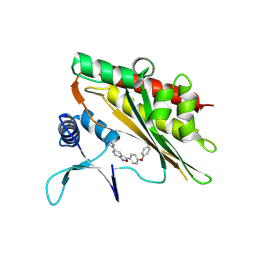 | |
8V7P
 
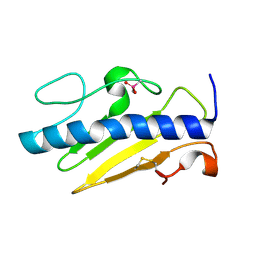 | |
7QTB
 
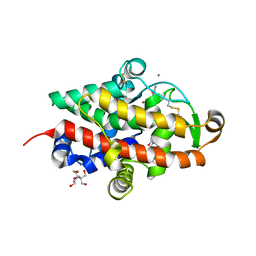 | | S1 nuclease from Aspergillus oryzae in complex with cytidine-5'-monophosphate | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Adamkova, K, Koval, T, Kolenko, P, Oestergaard, L.H, Dohnalek, J. | | Deposit date: | 2022-01-14 | | Release date: | 2022-11-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Atomic resolution studies of S1 nuclease complexes reveal details of RNA interaction with the enzyme despite multiple lattice-translocation defects.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
8TXN
 
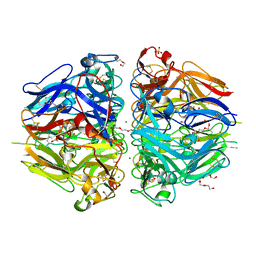 | |
8V74
 
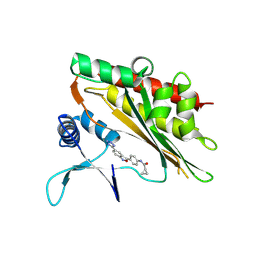 | |
7QTA
 
 | | S1 nuclease from Aspergillus oryzae in complex with uridine | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Nuclease S1, ... | | Authors: | Adamkova, K, Koval, T, Kolenko, P, Oestergaard, L.H, Dohnalek, J. | | Deposit date: | 2022-01-14 | | Release date: | 2022-11-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Atomic resolution studies of S1 nuclease complexes reveal details of RNA interaction with the enzyme despite multiple lattice-translocation defects.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
8V6Y
 
 | |
8V71
 
 | |
7QKN
 
 | | Crystal structure of YTHDF1 YTH domain in complex with the ebsulfur derivative compound 7 | | Descriptor: | 1,2-ETHANEDIOL, THIOCYANATE ION, YTH domain-containing family protein 1, ... | | Authors: | Dalle Vedove, A, Cazzanelli, G, Quattrone, A, Provenzani, A, Lolli, G. | | Deposit date: | 2021-12-18 | | Release date: | 2022-11-23 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Small-Molecule Ebselen Binds to YTHDF Proteins Interfering with the Recognition of N 6 -Methyladenosine-Modified RNAs.
Acs Pharmacol Transl Sci, 5, 2022
|
|
8V70
 
 | |
8V77
 
 | |
7Q9X
 
 | | Crystal structure of Chromobacterium violaceum aminotransferase in complex with PLP-pyruvate adduct | | Descriptor: | (3E)-4-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}-2-oxobut-3-enoic acid, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Isupov, M.N, Mitchell, D, Sayer, C, Littlechild, J.A. | | Deposit date: | 2021-11-15 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Aminotransferase from Chromobacterium violaceum in complex with PLP-pyruvate adduct.
To Be Published
|
|
8V6Z
 
 | |
7QRC
 
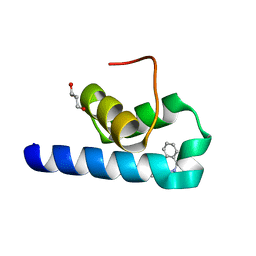 | | X-ray structure of Trypanosoma cruzi PEX14 in complex with a PEX5-PEX14 PPI inhibitor | | Descriptor: | GLYCEROL, Peroxin-14, ~{N}-(5-ethyl-6-oxidanylidene-benzo[b][1,4]benzothiazepin-2-yl)-2-(4-fluorophenyl)ethanamide | | Authors: | Napolitano, V, Popowicz, G.M, Dawidowski, M, Dubin, G. | | Deposit date: | 2022-01-10 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structure-based design, synthesis and evaluation of a novel family of PEX5-PEX14 interaction inhibitors against Trypanosoma.
Eur.J.Med.Chem., 243, 2022
|
|
8XWZ
 
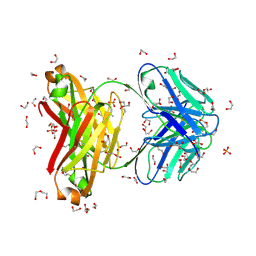 | | Non-crystalline proximal tubulopathy and crystalline cast nephropathy-causing Bence-Jones protein PT-CN: An entire immunoglobulin kappa light chain dimer | | Descriptor: | 1,2-ETHANEDIOL, 2-ETHOXYETHANOL, Bence-Jones protein kappa light chain PT-CN, ... | | Authors: | Ezawa, T, Otomo, R, Nozawa, K, Kyoya, S, Noguchi, K, Yohda, M, Odaka, M, Matsumura, H. | | Deposit date: | 2024-01-17 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Multiple myeloma-associated non-crystalline proximal tubulopathy and crystalline cast nephropathy: Biochemical and structural features of disease-causing monoclonal kappa light chains.
Faseb J., 39, 2025
|
|
8V72
 
 | |
8V76
 
 | |
7STR
 
 | | Crystal Structure of Human Fab S24-1063 in the Complex with the N-teminal Domain of Nucleocapsid Protein from SARS CoV-2 | | Descriptor: | 1,2-ETHANEDIOL, Fab S24-1063, Heavy chain, ... | | Authors: | Kim, Y, Maltseva, N, Tesar, C, Jedrzejczak, R, Dugan, H, Stamper, C, Wilson, P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-11-15 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Epitopes recognition of SARS-CoV-2 nucleocapsid RNA binding domain by human monoclonal antibodies.
Iscience, 27, 2024
|
|
7Q9Z
 
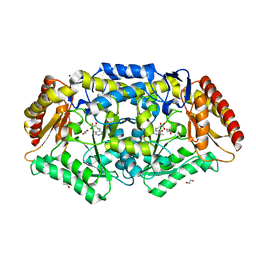 | | Crystal structure of Chromobacterium violaceum aminotransferase in complex with PLP-pyruvate adduct | | Descriptor: | (3E)-4-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}-2-oxobut-3-enoic acid, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Isupov, M.N, Mitchell, D, Sayer, C, Littlechild, J.A. | | Deposit date: | 2021-11-15 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Aminotransferase from Chromobacterium violaceum in complex with PLP-pyruvate adduct.
To Be Published
|
|
