7V64
 
 | | Crystal structure of Antibody 16A in complex with MUC1 Glycopeptide(GlycoT) | | Descriptor: | 16A fab Heavy chain, 16A fab Light chain, 2-acetamido-2-deoxy-beta-D-galactopyranose, ... | | Authors: | Niu, J, Xu, L, Meng, B, Han, Y.B, Yang, B. | | Deposit date: | 2021-08-19 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Site-specific GalNAc modification on a MUC1 neoantigen epitope forms a basis for high-affinity antibody binding
To Be Published
|
|
7V8Q
 
 | | Crystal structure of antibody 14A in complex with MUC1 Glycopeptide(GlycoT) | | Descriptor: | 14A fab heavy chain, 14A fab light chain, 2-acetamido-2-deoxy-beta-D-galactopyranose, ... | | Authors: | Niu, J, Xu, L, Meng, B, Han, Y.B, Yang, B. | | Deposit date: | 2021-08-23 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Site-specific GalNAc modification on a MUC1 neoantigen epitope forms a basis for high-affinity antibody binding
To Be Published
|
|
7VAC
 
 | | Crystal structure of antibody 14A in complex with MUC1 glycopeptide(GlycoST) | | Descriptor: | 14A fab heavy chain, 14A fab light chain, 2-acetamido-2-deoxy-beta-D-galactopyranose, ... | | Authors: | Niu, J, Xu, L, Meng, B, Han, Y.B, Yang, B. | | Deposit date: | 2021-08-28 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Site-specific GalNAc modification on a MUC1 neoantigen epitope forms a basis for high-affinity antibody binding
To Be Published
|
|
7VAZ
 
 | | Crystal structure of antibody 14A in complex with MUC1 glycopeptide(GlycoS) | | Descriptor: | 14A fab heavy chain, 14A fab light chain, 2-acetamido-2-deoxy-beta-D-galactopyranose, ... | | Authors: | Niu, J, Xu, L, Meng, B, Han, Y.B, Yang, B. | | Deposit date: | 2021-08-30 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Site-specific GalNAc modification on a MUC1 neoantigen epitope forms a basis for high-affinity antibody binding
To Be Published
|
|
7V6N
 
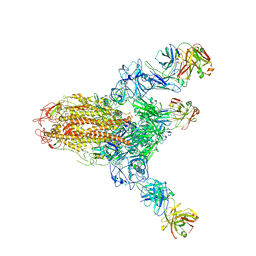 | | MERS S ectodomain trimer in complex with neutralizing antibody 111 state1 | | Descriptor: | 111 H, 111 L, Spike glycoprotein | | Authors: | Wang, X, Zhao, J, Wang, Z, Zeng, J, Zhang, S, Wang, Y. | | Deposit date: | 2021-08-20 | | Release date: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (3.99 Å) | | Cite: | MERS S ectodomain trimer in complex with neutralizing antibody 111 state1
to be published
|
|
3VTT
 
 | |
7VTJ
 
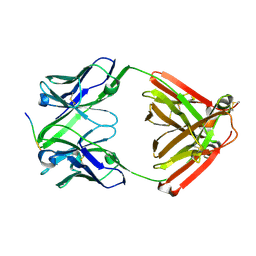 | | The cross-reaction complex structure with VQIIYK peptide and tau antibody's Fab domain. | | Descriptor: | Heavy chain of Fab, Light chain of Fab, VQIIYK peptide | | Authors: | Tsuchida, T, Fukuhara, N, Tsuchiya, T, Miyamoto, K, In, Y, Minoura, K, Taniguchi, Y, Ishida, T, Tomoo, K. | | Deposit date: | 2021-10-29 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The cross-reaction complex structure with VQIIYK peptide and tau antibody's Fab domain.
To Be Published
|
|
7V4W
 
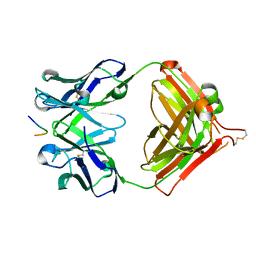 | | Crystal structure of Antibody 16A in complex with MUC1 peptide | | Descriptor: | 16A Fab Heavy chain, 16A Fab Light chain, Mucin-1 subunit alpha | | Authors: | Niu, J, Xu, L, Meng, B, Han, Y.B, Yang, B. | | Deposit date: | 2021-08-15 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Site-specific GalNAc modification on a MUC1 neoantigen epitope forms a basis for high-affinity antibody binding
To Be Published
|
|
7FG2
 
 | |
7FG3
 
 | |
7FG7
 
 | |
8RGZ
 
 | | Trimeric HSV-1F gB ectodomain in postfusion conformation with three bound HDIT101 Fab molecules. | | Descriptor: | Envelope glycoprotein B, HDIT101 Fab heavy chain, HDIT101 Fab light chain | | Authors: | Kalbermatter, D, Seyfizadeh, N, Imhof, T, Ries, M, Mueller, C, Jenner, L, Blumenschein, E, Yendrzheyevskiy, A, Moog, K, Eckert, D, Engel, R, Diebolder, P, Chami, M, Krauss, J, Schaller, T, Arndt, M. | | Deposit date: | 2023-12-14 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Development of a highly effective combination monoclonal antibody therapy against Herpes simplex virus.
J.Biomed.Sci., 31, 2024
|
|
8RH0
 
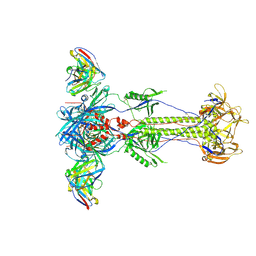 | | Trimeric HSV-1F gB ectodomain in postfusion conformation with three bound HDIT102 Fab molecules. | | Descriptor: | Envelope glycoprotein B, HDIT102 Fab heavy chain | | Authors: | Kalbermatter, D, Seyfizadeh, N, Imhof, T, Ries, M, Mueller, C, Jenner, L, Blumenschein, E, Yendrzheyevskiy, A, Moog, K, Eckert, D, Engel, R, Diebolder, P, Chami, M, Krauss, J, Schaller, T, Arndt, M. | | Deposit date: | 2023-12-14 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Development of a highly effective combination monoclonal antibody therapy against Herpes simplex virus.
J.Biomed.Sci., 31, 2024
|
|
8GHZ
 
 | | Cryo-EM structure of fish immunogloblin M-Fc | | Descriptor: | Teleost immunoglobulin M protein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Lyu, M, Stadtmueller, B.M, Malyutin, A.G. | | Deposit date: | 2023-03-13 | | Release date: | 2023-11-08 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | The structure of the teleost Immunoglobulin M core provides insights on polymeric antibody evolution, assembly, and function.
Nat Commun, 14, 2023
|
|
8G8N
 
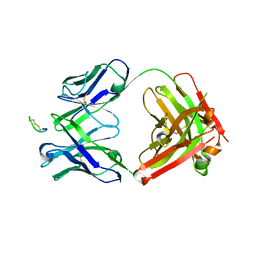 | | CTLA4 Fab with peptide | | Descriptor: | CYS-PRO-GLY-LYS-GLY-LEU-PRO-SER-CYS, Fab heavy chain, Fab light chain | | Authors: | Williams, J.C. | | Deposit date: | 2023-02-18 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | XTX101, a tumor-activated, Fc-enhanced anti-CTLA-4 monoclonal antibody, demonstrates tumor-growth inhibition and tumor-selective pharmacodynamics in mouse models of cancer.
J Immunother Cancer, 11, 2023
|
|
8SX3
 
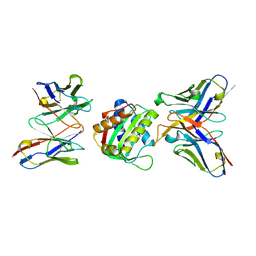 | | 10E8-GT10.2 immunogen in complex with human Fab 10E8 and mouse Fab W6-10 | | Descriptor: | 10E8 Fab heavy chain, 10E8 light chain, 10E8-GT10.2 immunogen, ... | | Authors: | Huang, J, Ozorowski, G, Ward, A.B. | | Deposit date: | 2023-05-19 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Vaccination induces broadly neutralizing antibody precursors to HIV gp41.
Nat.Immunol., 25, 2024
|
|
5XS5
 
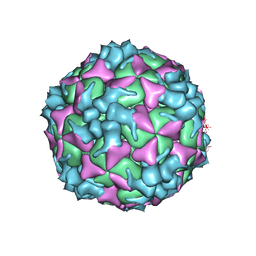 | | Structure of Coxsackievirus A6 (CVA6) virus procapsid particle | | Descriptor: | Genome polyprotein | | Authors: | Zheng, Q.B, He, M.Z, Xu, L.F, Yu, H, Cheng, T, Li, S.W. | | Deposit date: | 2017-06-12 | | Release date: | 2017-09-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Atomic structures of Coxsackievirus A6 and its complex with a neutralizing antibody
Nat Commun, 8, 2017
|
|
7UKM
 
 | |
8R80
 
 | | SARS-CoV-2 Delta RBD in complex with XBB-9 Fab and an anti-Fab nanobody | | Descriptor: | Spike protein S1, XBB-9 Fab heavy chain, XBB-9 Fab light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2023-11-27 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (4.03 Å) | | Cite: | A structure-function analysis shows SARS-CoV-2 BA.2.86 balances antibody escape and ACE2 affinity.
Cell Rep Med, 5, 2024
|
|
8RH1
 
 | | Trimeric HSV-2F gB ectodomain in postfusion conformation with three bound HDIT101 Fab molecules. | | Descriptor: | Envelope glycoprotein B, HDIT101 Fab heavy chain, HDIT101 Fab light chain | | Authors: | Kalbermatter, D, Seyfizadeh, N, Imhof, T, Ries, M, Mueller, C, Jenner, L, Blumenschein, E, Yendrzheyevskiy, A, Moog, K, Eckert, D, Engel, R, Diebolder, P, Chami, M, Krauss, J, Schaller, T, Arndt, M. | | Deposit date: | 2023-12-14 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Development of a highly effective combination monoclonal antibody therapy against Herpes simplex virus.
J.Biomed.Sci., 31, 2024
|
|
8RH2
 
 | | Trimeric HSV-2G gB ectodomain in postfusion conformation with three bound HDIT102 Fab molecules. | | Descriptor: | Envelope glycoprotein B, HDIT102 Fab heavy chain, HDIT102 Fab light chain | | Authors: | Kalbermatter, D, Seyfizadeh, N, Imhof, T, Ries, M, Mueller, C, Jenner, L, Blumenschein, E, Yendrzheyevskiy, A, Moog, K, Eckert, D, Engel, R, Diebolder, P, Chami, M, Krauss, J, Schaller, T, Arndt, M. | | Deposit date: | 2023-12-14 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Development of a highly effective combination monoclonal antibody therapy against Herpes simplex virus.
J.Biomed.Sci., 31, 2024
|
|
8QRF
 
 | | SARS-CoV-2 delta RBD complexed with XBB-6 and beta-49 Fabs | | Descriptor: | Beta-49 heavy chain, Beta-49 light chain, Spike protein S1, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2023-10-06 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | A structure-function analysis shows SARS-CoV-2 BA.2.86 balances antibody escape and ACE2 affinity.
Cell Rep Med, 5, 2024
|
|
8QTD
 
 | | Local refinement of SARS-CoV-2 BA.2.86 Spike and XBB-7 Fab | | Descriptor: | Spike glycoprotein,Fibritin, XBB-7 fab heavy chain, XBB-7 fab light chain | | Authors: | Ren, J, Duyvesteyn, H.M.E, Stuart, D.I. | | Deposit date: | 2023-10-12 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A structure-function analysis shows SARS-CoV-2 BA.2.86 balances antibody escape and ACE2 affinity.
Cell Rep Med, 5, 2024
|
|
8QRG
 
 | | SARS-CoV-2 delta RBD complexed with XBB-2 Fab and NbC1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, NbC1, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2023-10-07 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A structure-function analysis shows SARS-CoV-2 BA.2.86 balances antibody escape and ACE2 affinity.
Cell Rep Med, 5, 2024
|
|
8QSQ
 
 | |
