2MJ0
 
 | |
2MJ1
 
 | |
2MJ2
 
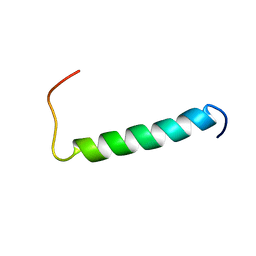 | | Structure of the dimerization domain of the human polyoma, JC virus agnoprotein is an amphipathic alpha-helix. | | Descriptor: | Agnoprotein | | Authors: | Coric, P, Saribas, S.A, Abou-Gharbia, M, Childers, W, White, M, Bouaziz, S, Safak, M. | | Deposit date: | 2013-12-23 | | Release date: | 2014-04-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The structure of the dimerization domain of the human polyoma, JC virus agnoprotein is an amphipathic alpha-helix
J.Virol., 2014
|
|
2MJ3
 
 | |
2MJ4
 
 | |
2MJ5
 
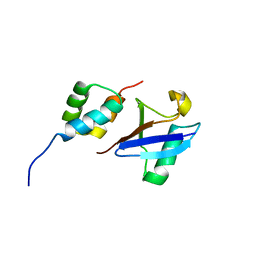 | | Structure of the UBA Domain of Human NBR1 in Complex with Ubiquitin | | Descriptor: | Next to BRCA1 gene 1 protein, Polyubiquitin-C | | Authors: | Walinda, E, Morimoto, D, Sugase, K, Komatsu, M, Tochio, H, Shirakawa, M. | | Deposit date: | 2013-12-25 | | Release date: | 2014-04-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the ubiquitin-associated (UBA) domain of human autophagy receptor NBR1 and its interaction with ubiquitin and polyubiquitin.
J.Biol.Chem., 289, 2014
|
|
2MJ6
 
 | |
2MJ7
 
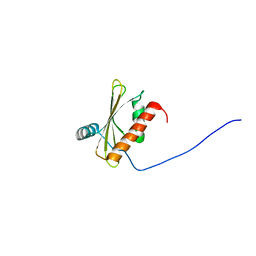 | | Solution NMR structure of beta-adaptin appendage domain of human adaptor protein complex 4 subunit beta, Northeast Structural Genomics Consortium (NESG) Target HR8998C | | Descriptor: | AP-4 complex subunit beta-1 | | Authors: | Eletsky, A, Rotshteyn, D.J, Pederson, K, Shastry, R, Maglaqui, M, Janjua, H, Xiao, R, Everett, J.K, Montelione, G.T, Prestegard, J.H, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-12-26 | | Release date: | 2014-01-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of beta-adaptin appendage domain of human adaptor protein complex 4 subunit beta, Northeast Structural Genomics Consortium (NESG) Target HR8998C
To be Published
|
|
2MJ8
 
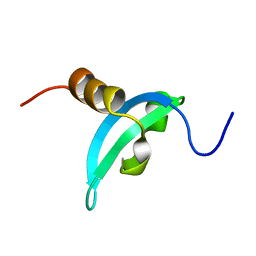 | | Solution structure of CDYL2 chromodomain | | Descriptor: | Chromodomain Y-like protein 2 | | Authors: | Qin, S, Houliston, S, Arrowsmith, C.H, Edwards, A.M, Wu, H, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-12-28 | | Release date: | 2014-04-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for the Binding Selectivity of Human CDY Chromodomains.
Cell Chem Biol, 27, 2020
|
|
2MJ9
 
 | | Designed Exendin-4 analogues | | Descriptor: | Exendin-4 | | Authors: | Rovo, P, Farkas, V, Straner, P, Szabo, M, Jermendy, A, Hegyi, O, Toth, G.K, Perczel, A. | | Deposit date: | 2013-12-30 | | Release date: | 2014-06-04 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Rational design of alpha-helix-stabilized exendin-4 analogues.
Biochemistry, 53, 2014
|
|
2MJA
 
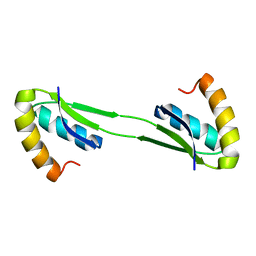 | |
2MJB
 
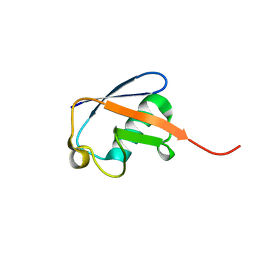 | | Solution nmr structure of ubiquitin refined against dipolar couplings in 4 media | | Descriptor: | Ubiquitin-60S ribosomal protein L40 | | Authors: | Maltsev, A, Grishaev, A, Roche, J, Zasloff, M, Bax, A. | | Deposit date: | 2014-01-02 | | Release date: | 2014-03-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Improved cross validation of a static ubiquitin structure derived from high precision residual dipolar couplings measured in a drug-based liquid crystalline phase.
J.Am.Chem.Soc., 136, 2014
|
|
2MJC
 
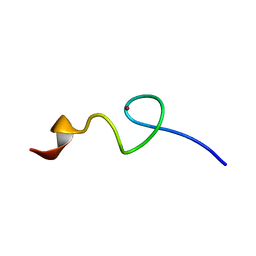 | | Zn-binding domain of eukaryotic translation initiation factor 3, subunit G | | Descriptor: | Eukaryotic translation initiation factor 3 subunit G, ZINC ION | | Authors: | Al-Abdul-Wahid, M, Menade, M, Xie, J, Kozlov, G, Gehring, K. | | Deposit date: | 2014-01-03 | | Release date: | 2015-01-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the Zn-binding domain of eukaryotic translation initiation factor 3, subunit G
To be Published
|
|
2MJD
 
 | | Oxidized Yeast Adrenodoxin Homolog 1 | | Descriptor: | Adrenodoxin homolog, mitochondrial, FE2/S2 (INORGANIC) CLUSTER | | Authors: | Gallo, A, Banci, L. | | Deposit date: | 2014-01-07 | | Release date: | 2014-11-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Functional reconstitution of mitochondrial Fe/S cluster synthesis on Isu1 reveals the involvement of ferredoxin.
Nat Commun, 5, 2014
|
|
2MJE
 
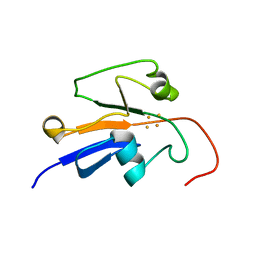 | | Reduced Yeast Adrenodoxin Homolog 1 | | Descriptor: | Adrenodoxin homolog, mitochondrial, FE2/S2 (INORGANIC) CLUSTER | | Authors: | Gallo, A, Banci, L. | | Deposit date: | 2014-01-07 | | Release date: | 2014-11-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Functional reconstitution of mitochondrial Fe/S cluster synthesis on Isu1 reveals the involvement of ferredoxin.
Nat Commun, 5, 2014
|
|
2MJF
 
 | | Solution structure of the complex between the yeast Rsa1 and Hit1 proteins | | Descriptor: | Protein HIT1, Ribosome assembly 1 protein | | Authors: | Quinternet, M, Roth, B, Back, R, Jacquemin, C, Manival, X. | | Deposit date: | 2014-01-08 | | Release date: | 2014-09-10 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Protein Hit1, a novel box C/D snoRNP assembly factor, controls cellular concentration of the scaffolding protein Rsa1 by direct interaction.
Nucleic Acids Res., 42, 2014
|
|
2MJG
 
 | |
2MJH
 
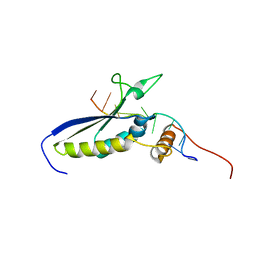 | |
2MJI
 
 | | HIFABP_Ketorolac_complex | | Descriptor: | (1R)-5-benzoyl-2,3-dihydro-1H-pyrrolizine-1-carboxylic acid, Fatty acid-binding protein, intestinal | | Authors: | Patil, R, Laguerre, A, Wielens, J, Headey, S, Williams, M, Mohanty, B, Porter, C, Scanlon, M. | | Deposit date: | 2014-01-09 | | Release date: | 2014-10-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Characterization of two distinct modes of drug binding to human intestinal Fatty Acid binding protein.
Acs Chem.Biol., 9, 2014
|
|
2MJJ
 
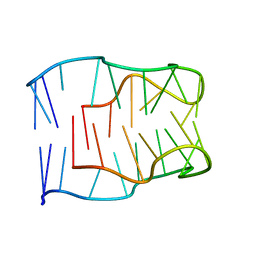 | |
2MJK
 
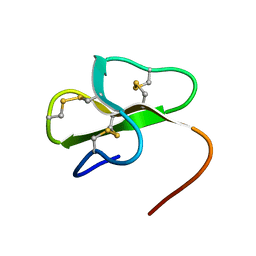 | |
2MJL
 
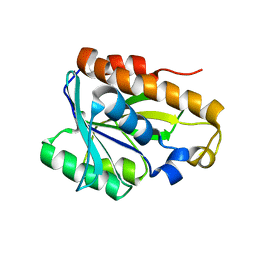 | | Solution structure of peptidyl-tRNA hyrolase from Vibrio cholerae | | Descriptor: | Peptidyl-tRNA hydrolase | | Authors: | Kabra, A, Shahid, S, Yadav, R, Pulavarti, S, Shukla, V.K, Arora, A. | | Deposit date: | 2014-01-11 | | Release date: | 2015-01-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of peptidyl-tRNA hydrolase from Vibrio cholerae
To be Published
|
|
2MJM
 
 | |
2MJN
 
 | |
2MJO
 
 | |
