1EUU
 
 | | SIALIDASE OR NEURAMINIDASE, LARGE 68KD FORM | | Descriptor: | SIALIDASE, SODIUM ION, beta-D-galactopyranose | | Authors: | Gaskell, A, Crennell, S.J, Taylor, G.L. | | Deposit date: | 1996-06-21 | | Release date: | 1997-01-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The three domains of a bacterial sialidase: a beta-propeller, an immunoglobulin module and a galactose-binding jelly-roll.
Structure, 3, 1995
|
|
1EQG
 
 | | THE 2.6 ANGSTROM MODEL OF OVINE COX-1 COMPLEXED WITH IBUPROFEN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IBUPROFEN, PROSTAGLANDIN H2 SYNTHASE-1, ... | | Authors: | Loll, P.J, Selinsky, B.S, Gupta, K, Sharkey, C.T. | | Deposit date: | 2000-04-04 | | Release date: | 2001-04-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural analysis of NSAID binding by prostaglandin H2 synthase: time-dependent and time-independent inhibitors elicit identical enzyme conformations.
Biochemistry, 40, 2001
|
|
1EFA
 
 | |
3MY7
 
 | | The Crystal Structure of the ACDH domain of an Alcohol Dehydrogenase from Vibrio parahaemolyticus to 2.25A | | Descriptor: | Alcohol dehydrogenase/acetaldehyde dehydrogenase, CHLORIDE ION | | Authors: | Stein, A.J, Weger, A, Volkart, L, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-05-10 | | Release date: | 2010-06-02 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal Structure of the ACDH domain of an Alcohol Dehydrogenase from Vibrio parahaemolyticus to 2.25A
To be Published
|
|
3MZY
 
 | |
6DOY
 
 | |
6DR1
 
 | | YopH PTP1B Chimera 2 PTPase | | Descriptor: | Targeted effector protein | | Authors: | Morales, Y, Johnson, S.J, Hengge, A.C. | | Deposit date: | 2018-06-11 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A YopH PTP1B Chimera Shows the Importance of the WPD-Loop Sequence to the Activity, Structure, and Dynamics of Protein Tyrosine Phosphatases.
Biochemistry, 57, 2018
|
|
6DR7
 
 | | YopH PTP1B WPD loop Chimera 2 PTPase bound to vanadate | | Descriptor: | ACETATE ION, Targeted effector protein, VANADATE ION | | Authors: | Moise, G, Morales, Y, Johnson, S.J, Hengge, A.C. | | Deposit date: | 2018-06-11 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | A YopH PTP1B Chimera Shows the Importance of the WPD-Loop Sequence to the Activity, Structure, and Dynamics of Protein Tyrosine Phosphatases.
Biochemistry, 57, 2018
|
|
3V8H
 
 | |
3MTI
 
 | |
7A4U
 
 | |
7AF0
 
 | | Structure of SARS-CoV-2 Main Protease bound to Ipidacrine. | | Descriptor: | 2,3,5,6,7,8-hexahydro-1~{H}-cyclopenta[b]quinolin-9-amine, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-09-18 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
6SBC
 
 | | Structure of type II terpene cyclase MstE from Scytonema in complex with farnesyl dihydroxybenzoate | | Descriptor: | CHLORIDE ION, MstE, SODIUM ION, ... | | Authors: | Moosmann, P, Ecker, F, Leopold-Messer, S, Cahn, J.K.B, Groll, M, Piel, J. | | Deposit date: | 2019-07-19 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A monodomain class II terpene cyclase assembles complex isoprenoid scaffolds.
Nat.Chem., 12, 2020
|
|
6S9S
 
 | | Dimerization domain of Xenopus laevis LDB1 in complex with darpin 10 | | Descriptor: | Darpin 10, LIM domain-binding protein 1 | | Authors: | Renko, M, Schaefer, J.V, Pluckthun, A, Bienz, M. | | Deposit date: | 2019-07-15 | | Release date: | 2019-10-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Rotational symmetry of the structured Chip/LDB-SSDP core module of the Wnt enhanceosome.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6SA0
 
 | |
6ZU8
 
 | | Crystal structure of human Brachyury G177D variant in complex with Afatinib | | Descriptor: | Brachyury protein, N-{4-[(3-chloro-4-fluorophenyl)amino]-7-[(3S)-tetrahydrofuran-3-yloxy]quinazolin-6-yl}-4-(dimethylamino)butanamide, ZINC ION | | Authors: | Newman, J.A, Gavard, A.E, Shrestha, L, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-07-21 | | Release date: | 2020-08-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of human Brachyury G177D variant in complex with Afatinib
To Be Published
|
|
5LJQ
 
 | | Crystal structure of human carbonic anhydrase II in complex with the 4-(4-(phenoxymethyl)-1H-1,2,3-triazol-1-yl)benzenesulfonamide inhibitor | | Descriptor: | 1-[4-[azanyl-bis(oxidanyl)-$l^{4}-sulfanyl]phenyl]-4-(phenoxymethyl)-1,2,3-triazole, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Ferraroni, M, Supuran, C. | | Deposit date: | 2016-07-19 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Benzenesulfonamides Incorporating Flexible Triazole Moieties Are Highly Effective Carbonic Anhydrase Inhibitors: Synthesis and Kinetic, Crystallographic, Computational, and Intraocular Pressure Lowering Investigations.
J. Med. Chem., 59, 2016
|
|
6SC6
 
 | | dAb3/HOIP-RBR apo structure | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase RNF31, SULFATE ION, ... | | Authors: | Tsai, Y.-C.I, House, D, Rittinger, K. | | Deposit date: | 2019-07-23 | | Release date: | 2019-11-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Single-Domain Antibodies as Crystallization Chaperones to Enable Structure-Based Inhibitor Development for RBR E3 Ubiquitin Ligases.
Cell Chem Biol, 27, 2020
|
|
6SBE
 
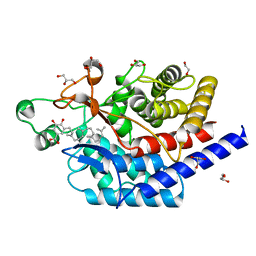 | | Structure of type II terpene cyclase MstE_D109N from Scytonema in complex with geranylgeranyl dihydroxybenzoate (substrate) | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, MstE, ... | | Authors: | Moosmann, P, Ecker, F, Leopold-Messer, S, Cahn, J.K.B, Groll, M, Piel, J. | | Deposit date: | 2019-07-19 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A monodomain class II terpene cyclase assembles complex isoprenoid scaffolds.
Nat.Chem., 12, 2020
|
|
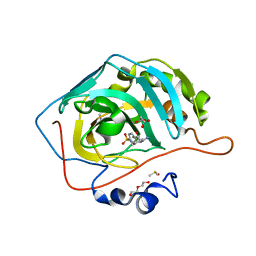
5LL4
 
 | |
6SCG
 
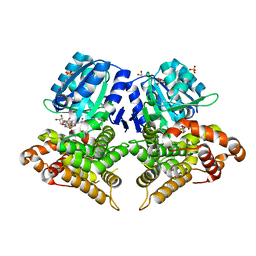 | | Structure of AdhE form 1 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Aldehyde-alcohol dehydrogenase, ... | | Authors: | Lovering, A.L, Bragginton, E. | | Deposit date: | 2019-07-24 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | High-resolution structure of the alcohol dehydrogenase domain of the bifunctional bacterial enzyme AdhE.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6SVL
 
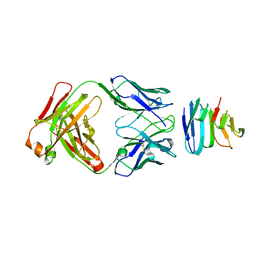 | |
5M2S
 
 | | R. flavefaciens' third ScaB cohesin in complex with a group 1 dockerin | | Descriptor: | CALCIUM ION, Doc8: Type I dockerin repeat domain from family 9 glycoside hydrolase WP_009982745[Ruminococcus flavefaciens], GLYCEROL, ... | | Authors: | Bule, P, Najmudin, S, Carvalho, A.L, Fontes, C.M.G.A. | | Deposit date: | 2016-10-13 | | Release date: | 2017-07-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Assembly of Ruminococcus flavefaciens cellulosome revealed by structures of two cohesin-dockerin complexes.
Sci Rep, 7, 2017
|
|
6SXP
 
 | |
6SYC
 
 | | Crystal structure of the lysozyme in presence of bromophenol blue at pH 6.5 | | Descriptor: | CHLORIDE ION, IMIDAZOLE, Lysozyme, ... | | Authors: | Camara-Artigas, A, Plaza-Garrido, M, Salinas-Garcia, M.C. | | Deposit date: | 2019-09-27 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Lysozyme crystals dyed with bromophenol blue: where has the dye gone?
Acta Crystallogr D Struct Biol, 76, 2020
|
|
