3A7F
 
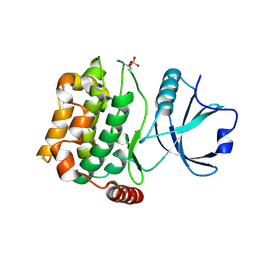 | | Human MST3 kinase | | Descriptor: | Serine/threonine kinase 24 (STE20 homolog, yeast) | | Authors: | Ko, T.P, Jeng, W.Y, Liu, C.I, Lai, M.D, Wang, A.H.J. | | Deposit date: | 2009-09-26 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structures of human MST3 kinase in complex with adenine, ADP and Mn2+.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3AGX
 
 | |
2ZYE
 
 | | Structure of HIV-1 Protease in Complex with Potent Inhibitor KNI-272 Determined by Neutron Crystallography | | Descriptor: | (4R)-N-tert-butyl-3-[(2S,3S)-2-hydroxy-3-({N-[(isoquinolin-5-yloxy)acetyl]-S-methyl-L-cysteinyl}amino)-4-phenylbutanoyl]-1,3-thiazolidine-4-carboxamide, protease | | Authors: | Adachi, M, Ohhara, T, Tamada, T, Okazaki, N, Kuroki, R. | | Deposit date: | 2009-01-20 | | Release date: | 2009-03-24 | | Last modified: | 2024-05-29 | | Method: | NEUTRON DIFFRACTION (1.9 Å) | | Cite: | Structure of HIV-1 protease in complex with potent inhibitor KNI-272 determined by high-resolution X-ray and neutron crystallography.
Proc.Natl.Acad.Sci.USA, 2009
|
|
3AO2
 
 | | Fragment-based approach to the design of ligands targeting a novel site on HIV-1 integrase | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 3-(7-bromo-1,3-benzodioxol-5-yl)-1-methyl-1H-pyrazol-5-amine, ... | | Authors: | Wielens, J, Chalmers, D.K, Headey, S.J, Parker, M.W, Scanlon, M.J. | | Deposit date: | 2010-09-20 | | Release date: | 2011-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fragment-based design of ligands targeting a novel site on the integrase enzyme of human immunodeficiency virus 1
Chemmedchem, 6, 2011
|
|
385D
 
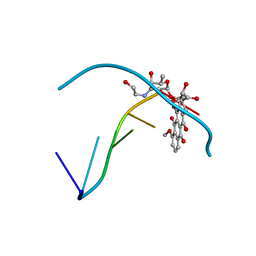 | |
397D
 
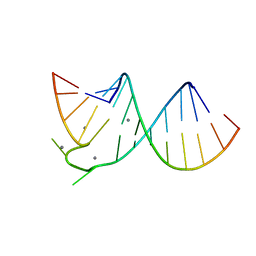 | |
362D
 
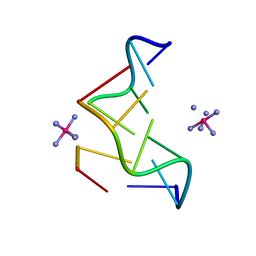 | | THE STRUCTURE OF D(TGCGCA)2 AND A COMPARISON TO OTHER Z-DNA HEXAMERS | | Descriptor: | COBALT HEXAMMINE(III), DNA (5'-D(*TP*GP*CP*GP*CP*A)-3') | | Authors: | Harper, N.A, Brannigan, J.A, Buck, M, Lewis, R.J, Moore, M.H, Schneider, B. | | Deposit date: | 1997-08-20 | | Release date: | 1997-11-20 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of d(TGCGCA)2 and a comparison to other DNA hexamers.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
3AAP
 
 | |
3AEX
 
 | |
3BQR
 
 | | Crystal structure of human death associated protein kinase 3 (DAPK3) in complex with an imidazo-pyridazine ligand | | Descriptor: | 4-(6-{[(1R)-1-(hydroxymethyl)propyl]amino}imidazo[1,2-b]pyridazin-3-yl)benzoic acid, Death-associated protein kinase 3, GLYCEROL, ... | | Authors: | Filippakopoulos, P, Rellos, P, Fedorov, O, Niesen, F, Pike, A.C.W, Pilka, E.S, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-12-20 | | Release date: | 2008-02-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of Human Death Associated Protein Kinase 3 (DAPK3) in Complex with an Imidazo-Pyridazine Ligand.
To be Published
|
|
3C4J
 
 | | ABC protein ArtP in complex with ATP-gamma-S | | Descriptor: | Amino acid ABC transporter (ArtP), MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Thaben, P.F, Eckey, V, Scheffel, F, Saenger, W, Schneider, E, Vahedi-Faridi, A. | | Deposit date: | 2008-01-30 | | Release date: | 2009-02-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal Structures of the ATP-binding cassette (ABC) Protein ArtP from Geobacillus stearothermophilus in complexes with nucleotides and nucleotide analogs reveal an intermediate semiclosed dimer in the post hydrolyses state and an asymmetry in the dimerisation region
To be Published
|
|
3C70
 
 | | HNL from Hevea brasiliensis to atomic resolution | | Descriptor: | Hydroxynitrilase, SULFATE ION, THIOCYANATE ION | | Authors: | Schmidt, A. | | Deposit date: | 2008-02-06 | | Release date: | 2008-06-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Atomic resolution crystal structures and quantum chemistry meet to reveal subtleties of hydroxynitrile lyase catalysis
J.Biol.Chem., 283, 2008
|
|
3C7Y
 
 | | Mutant R96A OF T4 lysozyme in wildtype background at 298K | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, lysozyme | | Authors: | Mooers, B.H.M. | | Deposit date: | 2008-02-08 | | Release date: | 2009-02-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Contributions of all 20 amino acids at site 96 to the stability and structure of T4 lysozyme.
Protein Sci., 18, 2009
|
|
3C3R
 
 | | ALIX BRO1 CHMP4C complex | | Descriptor: | Charged multivesicular body protein 4c peptide, GLYCEROL, Programmed cell death 6-interacting protein | | Authors: | McCullough, J.B, Fisher, R.D, Whitby, F.G, Sundquist, W.I, Hill, C.P. | | Deposit date: | 2008-01-28 | | Release date: | 2008-06-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | ALIX-CHMP4 interactions in the human ESCRT pathway.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3C82
 
 | | Bacteriophage lysozyme T4 lysozyme mutant K85A/R96H | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme | | Authors: | Mooers, B.H.M. | | Deposit date: | 2008-02-08 | | Release date: | 2009-02-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Contributions of all 20 amino acids at site 96 to the stability and structure of T4 lysozyme.
Protein Sci., 18, 2009
|
|
3BUS
 
 | | Crystal Structure of RebM | | Descriptor: | Methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | McCoy, J.G, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2008-01-03 | | Release date: | 2008-03-25 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure and mechanism of the rebeccamycin sugar 4'-O-methyltransferase RebM.
J.Biol.Chem., 283, 2008
|
|
3BX8
 
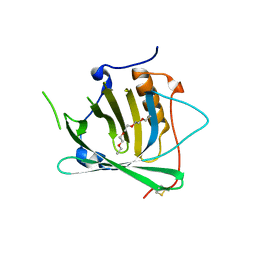 | | Engineered Human Lipocalin 2 (LCN2), apo-form | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ENGINEERED HUMAN LIPOCALIN 2, PENTAETHYLENE GLYCOL | | Authors: | Schonfeld, D.L, Chatwell, L, Skerra, A. | | Deposit date: | 2008-01-11 | | Release date: | 2009-01-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High affinity molecular recognition and functional blockade of CTLA-4 by an engineered human lipocalin
To be Published
|
|
3BWT
 
 | |
2ZUU
 
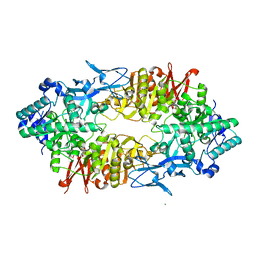 | | Crystal structure of Galacto-N-biose/Lacto-N-biose I phosphorylase in complex with GlcNAc | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, GLYCEROL, Lacto-N-biose phosphorylase, ... | | Authors: | Hidaka, M, Nishimoto, M, Kitaoka, M, Wakagi, T, Shoun, H, Fushinobu, S. | | Deposit date: | 2008-10-28 | | Release date: | 2008-12-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of galacto-N-biose/lacto-N-biose I phosphorylase: A large deformation of a tim barrel scaffold
J.Biol.Chem., 284, 2009
|
|
2Z45
 
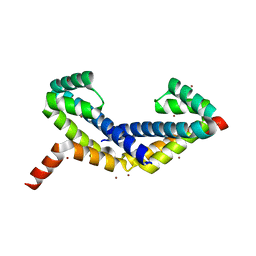 | | Crystal Structure of Zn-bound ORF134 | | Descriptor: | ORF134, ZINC ION | | Authors: | Tomimoto, Y, Ihara, K, Onizuka, T, Kanai, S, Ashida, H, Yokota, A, Tanaka, S, Miyasaka, H, Yamada, Y, Kato, R, Wakatsuki, S. | | Deposit date: | 2007-06-12 | | Release date: | 2008-06-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of ORF134
To be Published
|
|
2Z3F
 
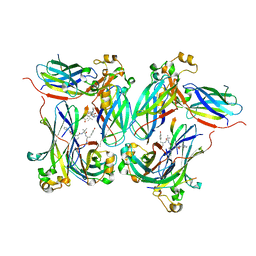 | |
2ZIR
 
 | | Crystal Structure of rat protein farnesyltransferase complexed with a benzofuran inhibitor and FPP | | Descriptor: | 2-[(S)-(4-chlorophenyl)(hydroxy)(1-methyl-1H-imidazol-5-yl)methyl]-N-morpholin-4-yl-7-phenyl-1-benzofuran-5-carboxamide, FARNESYL DIPHOSPHATE, GLYCEROL, ... | | Authors: | Fukami, T.A, Sogabe, S, Nagata, Y, Kondoh, O, Ishii, N. | | Deposit date: | 2008-02-22 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Synthesis and structure-activity relationships of novel benzofuran farnesyltransferase inhibitors
Bioorg.Med.Chem.Lett., 19, 2009
|
|
2Z4S
 
 | | Crystal structure of domain III from the Thermotoga maritima replication initiation protein DnaA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chromosomal replication initiator protein dnaA, MAGNESIUM ION | | Authors: | Fujikawa, N, Ozaki, S, Kagawa, W, Park, S.-Y, Katayama, T, Kurumizaka, H, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-06-25 | | Release date: | 2008-02-19 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A Common Mechanism for the ATP-DnaA-dependent Formation of Open Complexes at the Replication Origin.
J.Biol.Chem., 283, 2008
|
|
2Z55
 
 | |
2ZJS
 
 | | Crystal Structure of SecYE translocon from Thermus thermophilus with a Fab fragment | | Descriptor: | Fab56 (heavy chain), Fab56 (light chain), Preprotein translocase SecE subunit, ... | | Authors: | Tsukazaki, T, Mori, H, Fukai, S, Ishitani, R, Perederina, A, Vassylyev, D.G, Ito, K, Nureki, O. | | Deposit date: | 2008-03-08 | | Release date: | 2008-10-14 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Conformational transition of Sec machinery inferred from bacterial SecYE structures
Nature, 455, 2008
|
|
