2RFJ
 
 | | Crystal structure of the bromo domain 1 in human bromodomain containing protein, testis specific (BRDT) | | Descriptor: | Bromodomain testis-specific protein | | Authors: | Filippakopoulos, P, Salah, E, Savitsky, P, Keates, T, Parizotto, E, Elkins, J, Pike, A.C.W, Ugochukwu, E, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Sundstrom, M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-09-30 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Histone recognition and large-scale structural analysis of the human bromodomain family.
Cell(Cambridge,Mass.), 149, 2012
|
|
2R87
 
 | |
3QA2
 
 | | X-ray Structure of ketohexokinase in complex with a pyrimidopyrimidine analog 2 | | Descriptor: | Ketohexokinase, N~8~-(cyclopropylmethyl)-N~4~-(2-methylphenyl)-2-(piperazin-1-yl)pyrimido[5,4-d]pyrimidine-4,8-diamine, SULFATE ION | | Authors: | Abad, M.C. | | Deposit date: | 2011-01-10 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.519 Å) | | Cite: | Inhibitors of Ketohexokinase: Discovery of Pyrimidinopyrimidines with Specific Substitution that Complements the ATP-Binding Site.
ACS Med Chem Lett, 2, 2011
|
|
2RLC
 
 | |
2REJ
 
 | | ABC-transporter choline binding protein in unliganded semi-closed conformation | | Descriptor: | PUTATIVE GLYCINE BETAINE ABC TRANSPORTER PROTEIN | | Authors: | Oswald, C, Smits, S.H.J, Hoeing, M, Sohn-Boeser, L, Le Rudulier, D, Schmitt, L, Bremer, E. | | Deposit date: | 2007-09-26 | | Release date: | 2008-09-16 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of the choline/acetylcholine substrate-binding protein ChoX from Sinorhizobium meliloti in the liganded and unliganded-closed states.
J.Biol.Chem., 283, 2008
|
|
3QSB
 
 | | Structure of E. coli polIIIbeta with (Z)-5-(1-((4'-Fluorobiphenyl-4-yl)methoxyimino)butyl)-2,2-dimethyl-4,6-dioxocyclohexanecarbonitrile | | Descriptor: | (1R,5R)-5-{(1Z)-N-[(4'-fluorobiphenyl-4-yl)methoxy]butanimidoyl}-2,2-dimethyl-4,6-dioxocyclohexanecarbonitrile, DNA polymerase III subunit beta | | Authors: | Wijffels, G, Johnson, W.M, Oakley, A.J, Turner, K, Epa, V.C, Briscoe, S.J, Polley, M, Liepa, A.J, Hofmann, A, Buchardt, J, Christensen, C, Prosselkov, P, Dalrymple, B.P, Alewood, P.F, Jennings, P.A, Dixon, N.E, Winkler, D.A. | | Deposit date: | 2011-02-20 | | Release date: | 2011-06-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Binding inhibitors of the bacterial sliding clamp by design
J.Med.Chem., 54, 2011
|
|
3QFE
 
 | |
2RDE
 
 | | Crystal structure of VCA0042 complexed with c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Uncharacterized protein VCA0042 | | Authors: | Benach, J, Swaminathan, S.S, Tamayo, R, Seetharaman, J, Handelman, S, Forouhar, F, Neely, H, Camilli, A, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-09-22 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The structural basis of cyclic diguanylate signal transduction by PilZ domains.
Embo J., 26, 2007
|
|
3EJJ
 
 | | Structure of M-CSF bound to the first three domains of FMS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Colony stimulating factor-1, Macrophage colony-stimulating factor 1 receptor | | Authors: | Chen, X, Liu, H, Focia, P.J, Shim, A, He, X. | | Deposit date: | 2008-09-18 | | Release date: | 2008-12-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of macrophage colony stimulating factor bound to FMS: diverse signaling assemblies of class III receptor tyrosine kinases.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2REG
 
 | | ABC-transporter choline binding protein in complex with choline | | Descriptor: | CHOLINE ION, PUTATIVE GLYCINE BETAINE-BINDING ABC TRANSPORTER PROTEIN | | Authors: | Oswald, C, Schimtt, L, Smits, S.H.J, Hoeing, M, Sohn-Boeser, L, Bremer, E. | | Deposit date: | 2007-09-26 | | Release date: | 2008-09-16 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of the choline/acetylcholine substrate-binding protein ChoX from Sinorhizobium meliloti in the liganded and unliganded-closed states.
J.Biol.Chem., 283, 2008
|
|
3QI4
 
 | | Crystal structure of PDE9A(Q453E) in complex with IBMX | | Descriptor: | 3-ISOBUTYL-1-METHYLXANTHINE, High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, MAGNESIUM ION, ... | | Authors: | Hou, J, Xu, J, Liu, M, Zhao, R, Lou, H, Ke, H. | | Deposit date: | 2011-01-26 | | Release date: | 2011-04-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural asymmetry of phosphodiesterase-9, potential protonation of a glutamic Acid, and role of the invariant glutamine.
Plos One, 6, 2011
|
|
3QK7
 
 | | Crystal structure of putative Transcriptional regulator from Yersinia pestis biovar Microtus str. 91001 | | Descriptor: | Transcriptional regulators | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2011-01-31 | | Release date: | 2011-02-09 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of putative Transcriptional regulator from Yersinia pestis biovar Microtus str. 91001
To be Published
|
|
3QJP
 
 | |
3EKC
 
 | |
3EPE
 
 | |
3QMP
 
 | |
3ELK
 
 | | Crystal structure of putative transcriptional regulator TA0346 from Thermoplasma acidophilum | | Descriptor: | CHLORIDE ION, Putative transcriptional regulator TA0346 | | Authors: | Grantz Saskova, K, Chruszcz, M, Evdokimova, E, Egorova, O, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-09-22 | | Release date: | 2008-09-30 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of putative transcriptional regulator TA0346 from Thermoplasma acidophilum
To be Published
|
|
2RKB
 
 | | Serine dehydratase like-1 from human cancer cells | | Descriptor: | POTASSIUM ION, PYRIDOXAL-5'-PHOSPHATE, Serine dehydratase-like | | Authors: | Yamada, T, Komoto, J, Kasuya, T, Mori, H, Ogawa, H, Takusagawa, F. | | Deposit date: | 2007-10-16 | | Release date: | 2008-04-01 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A catalytic mechanism that explains a low catalytic activity of serine dehydratase like-1 from human cancer cells: Crystal structure and site-directed mutagenesis studies.
Biochim.Biophys.Acta, 1780, 2008
|
|
3QO1
 
 | | Monoclinic form of IgG1 Fab fragment (apo form) sharing same Fv as IgA | | Descriptor: | Fab fragment of IMMUNOGLOBULIN G1 HEAVY CHAIN, Fab fragment of IMMUNOGLOBULIN G1 LIGHT CHAIN, GLYCEROL | | Authors: | Trajtenberg, F, Correa, A, Buschiazzo, A. | | Deposit date: | 2011-02-09 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of a human IgA1 Fab fragment at 1.55 angstrom resolution: potential effect of the constant domains on antigen-affinity modulation
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3QUA
 
 | |
2RB9
 
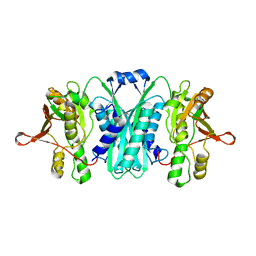 | | Crystal structure of E.coli HypE | | Descriptor: | HypE protein | | Authors: | Asinas, A.E, Rangarajan, E.S, Min, T, Matte, A, Proteau, A, Munger, C, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2007-09-18 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of [NiFe] hydrogenase maturation protein HypE from Escherichia coli and its interaction with HypF.
J.Bacteriol., 190, 2008
|
|
3QRE
 
 | |
3EY3
 
 | |
3QRS
 
 | |
2RFA
 
 | |
