9OK2
 
 | |
9SLY
 
 | |
9R70
 
 | |
9O0K
 
 | | ChtA CR domain from Corynebacterium diphtheriae | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Ford, J, Sawaya, M.R, Clubb, R.T. | | Deposit date: | 2025-04-02 | | Release date: | 2025-09-03 | | Last modified: | 2025-10-08 | | Method: | X-RAY DIFFRACTION (1.629 Å) | | Cite: | Structural basis of heme scavenging by the ChtA and HtaA hemophores in Corynebacterium diphtheriae.
J.Biol.Chem., 301, 2025
|
|
9R6Y
 
 | |
9R74
 
 | |
1EVK
 
 | | CRYSTAL STRUCTURE OF A TRUNCATED FORM OF THREONYL-TRNA SYNTHETASE WITH THE LIGAND THREONINE | | Descriptor: | THREONINE, THREONYL-TRNA SYNTHETASE, ZINC ION | | Authors: | Sankaranarayanan, R, Dock-Bregeon, A.C, Rees, B, Moras, D. | | Deposit date: | 2000-04-20 | | Release date: | 2000-07-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Zinc ion mediated amino acid discrimination by threonyl-tRNA synthetase.
Nat.Struct.Biol., 7, 2000
|
|
102D
 
 | |
1EKF
 
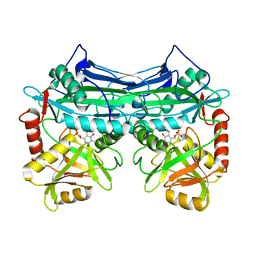 | | CRYSTALLOGRAPHIC STRUCTURE OF HUMAN BRANCHED CHAIN AMINO ACID AMINOTRANSFERASE (MITOCHONDRIAL) COMPLEXED WITH PYRIDOXAL-5'-PHOSPHATE AT 1.95 ANGSTROMS (ORTHORHOMBIC FORM) | | Descriptor: | BRANCHED CHAIN AMINO ACID AMINOTRANSFERASE (MITOCHONDRIAL), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Yennawar, N.H, Dunbar, J.H, Conway, M, Hutson, S.M, Farber, G.K. | | Deposit date: | 2000-03-08 | | Release date: | 2001-03-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The structure of human mitochondrial branched-chain aminotransferase.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1EWQ
 
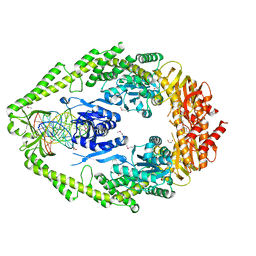 | | CRYSTAL STRUCTURE TAQ MUTS COMPLEXED WITH A HETERODUPLEX DNA AT 2.2 A RESOLUTION | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*CP*GP*AP*CP*GP*CP*TP*AP*GP*CP*GP*TP*GP*CP*GP*GP*CP*TP*CP*GP*TP*C)-3'), DNA (5'-D(*GP*GP*AP*CP*GP*AP*GP*CP*CP*GP*CP*CP*GP*CP*TP*AP*GP*CP*GP*TP*CP*G)-3'), ... | | Authors: | Obmolova, G, Ban, C, Hsieh, P, Yang, W. | | Deposit date: | 2000-04-26 | | Release date: | 2000-10-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of mismatch repair protein MutS and its complex with a substrate DNA.
Nature, 407, 2000
|
|
7OCJ
 
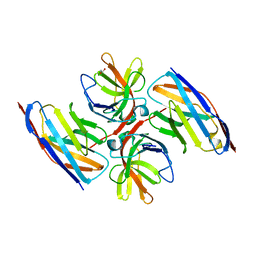 | | Crystal structure of E.coli LexA in complex with nanobody NbSOS2(Nb14509) | | Descriptor: | 1,2-ETHANEDIOL, LexA repressor, NbSOS2 (14509) | | Authors: | Maso, L, Vascon, F, Chinellato, M, Pardon, E, Steyaert, J, Angelini, A, Tondi, D, Cendron, L. | | Deposit date: | 2021-04-27 | | Release date: | 2022-10-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Nanobodies targeting LexA autocleavage disclose a novel suppression strategy of SOS-response pathway.
Structure, 30, 2022
|
|
8EYU
 
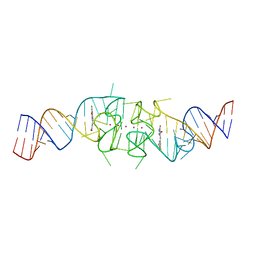 | | Structure of Beetroot dimer bound to DFAME | | Descriptor: | POTASSIUM ION, RNA (49-MER), methyl (2E)-3-{(4Z)-4-[(3,5-difluoro-4-hydroxyphenyl)methylidene]-1-methyl-5-oxo-4,5-dihydro-1H-imidazol-2-yl}prop-2-enoate | | Authors: | Passalacqua, L.F.M, Ferre-D'Amare, A.R. | | Deposit date: | 2022-10-28 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Co-crystal structures of the fluorogenic aptamer Beetroot show that close homology may not predict similar RNA architecture.
Nat Commun, 14, 2023
|
|
1F5S
 
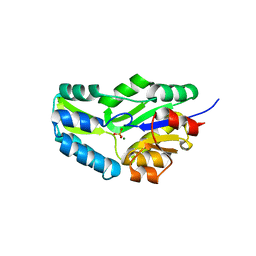 | | CRYSTAL STRUCTURE OF PHOSPHOSERINE PHOSPHATASE FROM METHANOCOCCUS JANNASCHII | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, PHOSPHOSERINE PHOSPHATASE (PSP) | | Authors: | Wang, W, Kim, R, Jancarik, J, Yokota, H, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2000-06-15 | | Release date: | 2001-06-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of phosphoserine phosphatase from Methanococcus jannaschii, a hyperthermophile, at 1.8 A resolution.
Structure, 9, 2001
|
|
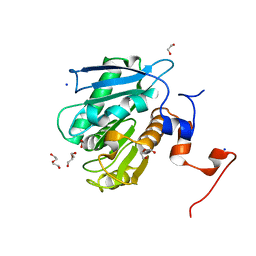
8EU1
 
 | |
5V2G
 
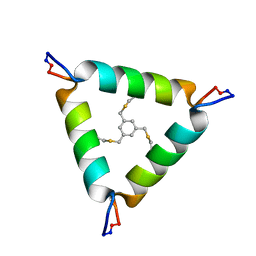 | | De Novo Design of Novel Covalent Constrained Meso-size Peptide Scaffolds with Unique Tertiary Structures | | Descriptor: | 1,3,5-tris(bromomethyl)benzene, 20-mer Peptide | | Authors: | Dang, B, Wu, H, Mulligan, V.K, Mravic, M, Wu, Y, Lemmin, T, Ford, A, Silva, D, Baker, D, DeGrado, W.F. | | Deposit date: | 2017-03-03 | | Release date: | 2017-09-27 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | De novo design of covalently constrained mesosize protein scaffolds with unique tertiary structures.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6NZX
 
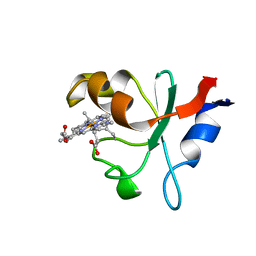 | | Hadesarchaea YNP_N21 cytochrome b5 domain protein (KUO41884.1) | | Descriptor: | Cytochrome B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Teakel, S.L, Marama, M.S, Aragao, D, Forwood, J.K, Cahill, M.A. | | Deposit date: | 2019-02-14 | | Release date: | 2019-03-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Hadesarchaea YNP_N21 cytochrome b5 domain protein (KUO41884.1)
To Be Published
|
|
1F9G
 
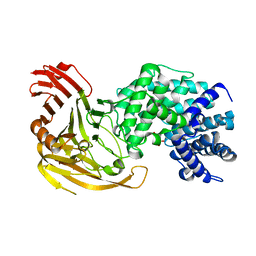 | |
1FH8
 
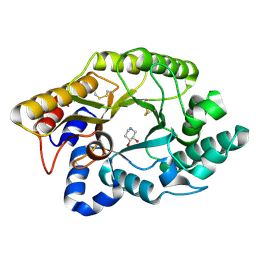 | | CRYSTAL STRUCTURE OF THE XYLANASE CEX WITH XYLOBIOSE-DERIVED ISOFAGOMINE INHIBITOR | | Descriptor: | BETA-1,4-XYLANASE, PIPERIDINE-3,4-DIOL, beta-D-xylopyranose | | Authors: | Notenboom, V, Williams, S.J, Hoos, R, Withers, S.G, Rose, D.R. | | Deposit date: | 2000-07-31 | | Release date: | 2000-08-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Detailed structural analysis of glycosidase/inhibitor interactions: complexes of Cex from Cellulomonas fimi with xylobiose-derived aza-sugars.
Biochemistry, 39, 2000
|
|
166D
 
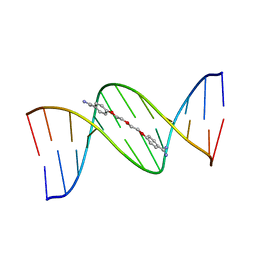 | |
6MVT
 
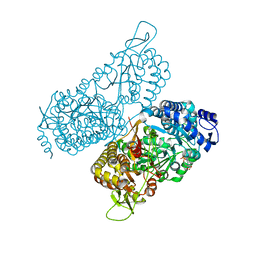 | | Structure of a bacterial ALDH16 complexed with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Aldehyde dehydrogenase, SODIUM ION | | Authors: | Tanner, J.J, Liu, L. | | Deposit date: | 2018-10-28 | | Release date: | 2018-12-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Aldehyde Dehydrogenase 16 Reveals Trans-Hierarchical Structural Similarity and a New Dimer.
J. Mol. Biol., 431, 2019
|
|
7P3M
 
 | |
7S6E
 
 | | Crystal structure of UrtA from Synechococcus CC9311 in complex with urea and calcium | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Shah, B.S, Mikolajek, H, Mykhaylyk, V, Orr, C.M, Owens, R.J, Paulsen, I.T. | | Deposit date: | 2021-09-14 | | Release date: | 2021-10-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.973 Å) | | Cite: | Crystal structure of UrtA from Synechococcus CC9311 in complex with urea and calcium
To Be Published
|
|
8X8P
 
 | | Phenylethanol rhamnosyltransferase (CmGT3) | | Descriptor: | 1,2-ETHANEDIOL, Phenylethanol rhamnosyltransferase (CmGT3) | | Authors: | Wang, H.T, Wang, Z.L, Ye, M. | | Deposit date: | 2023-11-28 | | Release date: | 2024-04-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Construct Phenylethanoid Glycosides Harnessing Biosynthetic Networks, Protein Engineering and One-Pot Multienzyme Cascades.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
1C9Z
 
 | | D232-CGTACG | | Descriptor: | 1,3-DI[[[10-METHOXY-7H-PYRIDO[4,3-C]CARBAZOL-2-IUMYL]-ETHYL]-PIPERIDIN-4-YL]-PROPANE, 5'-D(*CP*GP*TP*AP*CP*G)-3' | | Authors: | Williams, L.D. | | Deposit date: | 1999-08-03 | | Release date: | 2000-04-02 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Effects of cationic charge on three-dimensional structures of intercalative complexes: structure of a bis-intercalated DNA complex solved by MAD phasing.
Curr.Med.Chem., 7, 2000
|
|
9OW2
 
 | | Crystal Structure of the Surface Protein (CD630_07380) from Clostridium difficile Strain 630 | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, CHLORIDE ION, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2025-06-02 | | Release date: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of the Surface Protein (CD630_07380) from Clostridium difficile Strain 630.
To Be Published
|
|
