5A2C
 
 | | Crystal Structure of Anoxybacillus Alpha-amylase Provides Insights into a New Glycosyl Hydrolase Subclass | | Descriptor: | ALPHA-AMYLASE, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Ng, C.L, Chai, K.P, Othman, N.F, Teh, A.H, Ho, K.L, Chan, K.G, Goh, K.M. | | Deposit date: | 2015-05-17 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Anoxybacillus Alpha-Amylase Provides Insights Into Maltose Binding of a New Glycosyl Hydrolase Subclass.
Sci.Rep., 6, 2016
|
|
5A2A
 
 | | Crystal Structure of Anoxybacillus Alpha-amylase Provides Insights into a New Glycosyl Hydrolase Subclass | | Descriptor: | ACETATE ION, APO FORM OF ANOXYBACILLUS ALPHA-AMYLASES, CALCIUM ION | | Authors: | Ng, C.L, Chai, K.P, Othman, N.F, Teh, A.H, Ho, K.L, Chan, K.G, Goh, K.M. | | Deposit date: | 2015-05-16 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Anoxybacillus Alpha-Amylase Provides Insights Into Maltose Binding of a New Glycosyl Hydrolase Subclass.
Sci.Rep., 6, 2016
|
|
5A2B
 
 | | Crystal Structure of Anoxybacillus Alpha-amylase Provides Insights into a New Glycosyl Hydrolase Subclass | | Descriptor: | ANOXYBACILLUS ALPHA-AMYLASE, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Ng, C.L, Chai, K.P, Othman, N.F, Teh, A.H, Ho, K.L, Chan, K.G, Goh, K.M. | | Deposit date: | 2015-05-17 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of Anoxybacillus Alpha-Amylase Provides Insights Into Maltose Binding of a New Glycosyl Hydrolase Subclass.
Sci.Rep., 6, 2016
|
|
5EMY
 
 | | Human Pancreatic Alpha-Amylase in complex with the mechanism based inactivator glucosyl epi-cyclophellitol | | Descriptor: | (1R,2R,3S,5R,6S)-2,3,5-trihydroxy-6-(hydroxymethyl)cyclohexyl alpha-D-glucopyranoside, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Caner, S, Brayer, G.D. | | Deposit date: | 2015-11-06 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.231 Å) | | Cite: | Glucosyl epi-cyclophellitol allows mechanism-based inactivation and structural analysis of human pancreatic alpha-amylase.
Febs Lett., 590, 2016
|
|
5E0F
 
 | | Human pancreatic alpha-amylase in complex with mini-montbretin A | | Descriptor: | 5,7-dihydroxy-4-oxo-2-(3,4,5-trihydroxyphenyl)-4H-chromen-3-yl 6-deoxy-2-O-{6-O-[(2E)-3-(3,4-dihydroxyphenyl)prop-2-enoyl]-beta-D-glucopyranosyl}-alpha-L-mannopyranoside, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Caner, S, Brayer, G.D. | | Deposit date: | 2015-09-28 | | Release date: | 2016-09-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Human pancreatic alpha-amylase in complex with mini-montbretin A
To Be Published
|
|
5TD4
 
 | |
5GQU
 
 | |
5GQV
 
 | |
5GQX
 
 | |
5GQW
 
 | |
5KEZ
 
 | |
5JY7
 
 | |
5N7J
 
 | |
5M99
 
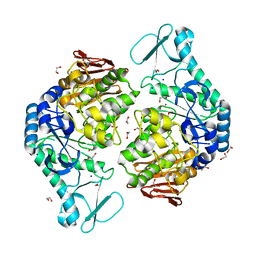 | | Functional Characterization and Crystal Structure of Thermostable Amylase from Thermotoga petrophila, reveals High Thermostability and an Archaic form of Dimerization | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha-amylase, ... | | Authors: | Hameed, U, Price, I, Mirza, O.A. | | Deposit date: | 2016-11-01 | | Release date: | 2017-07-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Functional characterization and crystal structure of thermostable amylase from Thermotoga petrophila, reveals high thermostability and an unusual form of dimerization.
Biochim. Biophys. Acta, 1865, 2017
|
|
5GR6
 
 | |
5GR4
 
 | |
5GQY
 
 | |
5GR1
 
 | |
5GR2
 
 | |
5GR5
 
 | |
5GQZ
 
 | |
5GR3
 
 | |
5GR0
 
 | |
5GTW
 
 | |
5H2T
 
 | |
