6X4F
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase (Y181C) Variant in Complex with methyl 2-(6-cyano-3-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-4-methylnaphthalen-1-yl)acetate (JLJ681), a Non-nucleoside Inhibitor | | Descriptor: | Reverse transcriptase/ribonuclease H, SULFATE ION, methyl (6-cyano-3-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}-4-methylnaphthalen-1-yl)acetate, ... | | Authors: | Chan, A.H, Duong, V.N, Ippolito, J.A, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2020-05-22 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structural investigation of 2-naphthyl phenyl ether inhibitors bound to WT and Y181C reverse transcriptase highlights key features of the NNRTI binding site.
Protein Sci., 29, 2020
|
|
6O48
 
 | | Wild-type HIV-1 protease in complex with a substrate analog CA-p2 | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Wang, Y.-F, Liu, F, Weber, I.T. | | Deposit date: | 2019-02-28 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Highly Drug-Resistant HIV-1 Protease Mutant PRS17 Shows Enhanced Binding to Substrate Analogues.
Acs Omega, 4, 2019
|
|
8B8B
 
 | | Multimerization domain of Munia virus 1 phosphoprotein | | Descriptor: | Munia Bornavirus 1 phosphoprotein, NITRATE ION | | Authors: | Chenavier, F, Tarbouriech, N, Bourhis, J.M, Tomonaga, K, Horie, M, Crepin, T. | | Deposit date: | 2022-10-04 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Borna Disease Virus 1 Phosphoprotein Forms a Tetramer and Interacts with Host Factors Involved in DNA Double-Strand Break Repair and mRNA Processing.
Viruses, 14, 2022
|
|
5J49
 
 | |
7RDK
 
 | | Crystal structure of PCDN-16B, an anti-HIV antibody from the PCDN bnAb lineage (cysteinylated state) | | Descriptor: | 1,2-ETHANEDIOL, CYSTEINE, GLYCEROL, ... | | Authors: | Omorodion, O, Wilson, I.A. | | Deposit date: | 2021-07-09 | | Release date: | 2021-11-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structural and Biochemical Characterization of Cysteinylation in Broadly Neutralizing Antibodies to HIV-1.
J.Mol.Biol., 433, 2021
|
|
7RB1
 
 | | Isocitrate Lyase-1 from Mycobacterium tuberculosis covalently modified by 5-descarboxy-5-nitro-D-isocitric acid | | Descriptor: | (3E)-3-(hydroxyimino)propanoic acid, GLYCEROL, GLYOXYLIC ACID, ... | | Authors: | Krieger, I.V, Mellott, D, Meek, T, Sacchettini, J.C. | | Deposit date: | 2021-07-05 | | Release date: | 2021-12-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism-Based Inactivation of Mycobacterium tuberculosis Isocitrate Lyase 1 by (2 R ,3 S )-2-Hydroxy-3-(nitromethyl)succinic acid.
J.Am.Chem.Soc., 143, 2021
|
|
6X3P
 
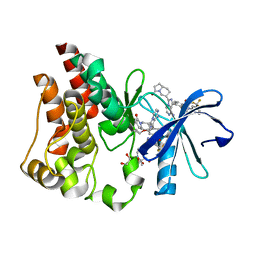 | | Co-structure of BTK kinase domain with L-005298385 inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-{8-amino-3-[(6R,8aS)-3-oxooctahydroindolizin-6-yl]imidazo[1,5-a]pyrazin-1-yl}-3-(cyclopropyloxy)-N-[4-(trifluoromethyl)pyridin-2-yl]benzamide, ... | | Authors: | Fischmann, T.O. | | Deposit date: | 2020-05-21 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Potent, non-covalent reversible BTK inhibitors with 8-amino-imidazo[1,5-a]pyrazine core featuring 3-position bicyclic ring substitutes.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
8G3Y
 
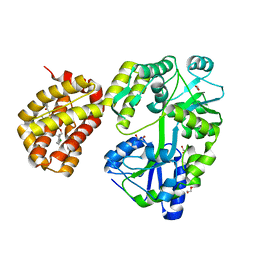 | | MBP-Mcl1 in complex with ligand 34 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Miller, B.R, Shaffer, P. | | Deposit date: | 2023-02-08 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of an Oral, Beyond-Rule-of-Five Mcl-1 Protein-Protein Interaction Modulator with the Potential of Treating Hematological Malignancies.
J.Med.Chem., 66, 2023
|
|
8G3X
 
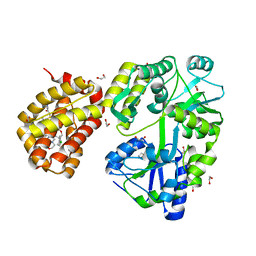 | | MBP-Mcl1 in complex with ligand 32 | | Descriptor: | 1,2-ETHANEDIOL, Maltodextrin-binding protein, Induced myeloid leukemia cell differentiation protein Mcl-1 chimera, ... | | Authors: | Miller, B.R, Shaffer, P. | | Deposit date: | 2023-02-08 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Discovery of an Oral, Beyond-Rule-of-Five Mcl-1 Protein-Protein Interaction Modulator with the Potential of Treating Hematological Malignancies.
J.Med.Chem., 66, 2023
|
|
6PBT
 
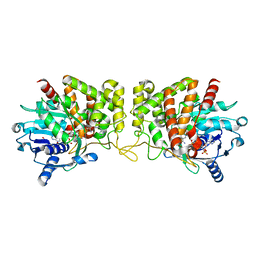 | | Pseudopaline Dehydrogenase with (R)-Pseudopaline Soaked 2 hours | | Descriptor: | 1,2-ETHANEDIOL, N-[(3S)-3-amino-3-carboxypropyl]-L-histidine, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | McFarlane, J.S, Lamb, A.L. | | Deposit date: | 2019-06-14 | | Release date: | 2019-10-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Staphylopine and pseudopaline dehydrogenase from bacterial pathogens catalyze reversible reactions and produce stereospecific metallophores.
J.Biol.Chem., 294, 2019
|
|
7R9P
 
 | | Crystal structure of HPK1 in complex with compound 14 | | Descriptor: | 6-amino-2-fluoro-N,N-dimethyl-3-(4'-methylspiro[cyclopropane-1,3'-pyrrolo[2,3-b]pyridin]-5'-yl)benzamide, Hematopoietic progenitor kinase, SULFATE ION | | Authors: | Wu, P, Lehoux, I, Wang, W. | | Deposit date: | 2021-06-29 | | Release date: | 2022-01-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Discovery of Spiro-azaindoline Inhibitors of Hematopoietic Progenitor Kinase 1 (HPK1).
Acs Med.Chem.Lett., 13, 2022
|
|
6ZFQ
 
 | | Structure of the catalytic domain of human endo-alpha-mannosidase MANEA in complex with bis-tris | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glycoprotein endo-alpha-1,2-mannosidase | | Authors: | Sobala, L.F, Fernandes, P.Z, Hakki, Z, Thompson, A.J, Howe, J.D, Hill, M, Zitzmann, N, Davies, S, Stamataki, Z, Butters, T.D, Alonzi, D.S, Williams, S.J, Davies, G.J. | | Deposit date: | 2020-06-17 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of human endo-alpha-1,2-mannosidase (MANEA), an antiviral host-glycosylation target.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3FIQ
 
 | | Odorant Binding Protein OBP1 | | Descriptor: | 1,2-ETHANEDIOL, Odorant-binding protein 1F | | Authors: | White, S.A. | | Deposit date: | 2008-12-12 | | Release date: | 2009-05-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Structure of rat odorant-binding protein OBP1 at 1.6 A resolution
Acta Crystallogr.,Sect.D, 65, 2009
|
|
8URJ
 
 | |
8G4T
 
 | | Vaccine-elicited human antibody 2C09 in complex with HIV-1 envelope trimer BG505 DS-SOSIP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein BG505 DS-SOSIP gp120, ... | | Authors: | Wang, S, Kwong, P.D. | | Deposit date: | 2023-02-10 | | Release date: | 2023-07-12 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | HIV-1 neutralizing antibodies elicited in humans by a prefusion-stabilized envelope trimer form a reproducible class targeting fusion peptide.
Cell Rep, 42, 2023
|
|
5KGT
 
 | | Crystal structure of 7,8-diaminopelargonic acid synthase (BioA) from Mycobacterium tuberculosis, complexed with an inhibitor optimized from HTS lead: 1-[4-[4-(3-chlorophenyl)carbonylpiperidin-1-yl]phenyl]ethanone | | Descriptor: | 1-[4-[4-(3-chlorophenyl)carbonylpiperidin-1-yl]phenyl]ethanone, Adenosylmethionine-8-amino-7-oxononanoate aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Maize, K.M, Divakaran, A, Aldrich, C.C, Finzel, B.C. | | Deposit date: | 2016-06-13 | | Release date: | 2017-06-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-Based Optimization of Pyridoxal 5'-Phosphate-Dependent Transaminase Enzyme (BioA) Inhibitors that Target Biotin Biosynthesis in Mycobacterium tuberculosis.
J. Med. Chem., 60, 2017
|
|
6O2L
 
 | |
6Z5V
 
 | | CRYSTAL STRUCTURE OF RAT PEROXISOMAL MULTIFUNCTIONAL ENZYME TYPE-1 (RPMFE1) COMPLEXED WITH 3-KETODECANOYL-COA IN CROTONASE FOLD AND OXIDISED NICOTINAMIDE ADENINE DINUCLEOTIDE IN HAD FOLD | | Descriptor: | 3-KETO-DECANOYL-COA, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Wierenga, R.K, Sridhar, S, Kiema, T.R. | | Deposit date: | 2020-05-27 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystallographic binding studies of rat peroxisomal multifunctional enzyme type 1 with 3-ketodecanoyl-CoA: capturing active and inactive states of its hydratase and dehydrogenase catalytic sites.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6Z5O
 
 | | CRYSTAL STRUCTURE OF RAT PEROXISOMAL MULTIFUNCTIONAL ENZYME TYPE-1 (RPMFE1) COMPLEXED WITH COENZYME-A AND OXIDISED NICOTINAMIDE ADENINE DINUCLEOTIDE | | Descriptor: | COENZYME A, GLYCEROL, NICOTINAMIDE, ... | | Authors: | Wierenga, R.K, Sridhar, S, Kiema, T.R. | | Deposit date: | 2020-05-27 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystallographic binding studies of rat peroxisomal multifunctional enzyme type 1 with 3-ketodecanoyl-CoA: capturing active and inactive states of its hydratase and dehydrogenase catalytic sites.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6ZDF
 
 | | Structure of the catalytic domain of human endo-alpha-mannosidase MANEA in complex with HEPES | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Glycoprotein endo-alpha-1,2-mannosidase | | Authors: | Sobala, L.F, Fernandes, P.Z, Hakki, Z, Thompson, A.J, Howe, J.D, Hill, M, Zitzmann, N, Davies, S, Stamataki, Z, Butters, T.D, Alonzi, D.S, Williams, S.J, Davies, G.J. | | Deposit date: | 2020-06-14 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of human endo-alpha-1,2-mannosidase (MANEA), an antiviral host-glycosylation target.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
9BCG
 
 | | Myeloid cell leukemia-1 (Mcl-1) complexed with compound | | Descriptor: | 7-[(4R,5S,6P)-7-chloro-10-[3-(4-chloro-3,5-dimethylphenoxy)propyl]-4-methyl-1-oxo-6-(1,3,5-trimethyl-1H-pyrazol-4-yl)-3,4-dihydropyrazino[1,2-a]indol-2(1H)-yl]-4,5-dimethoxy-1-methyl-1H-indole-2-carboxylic acid, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Zhao, B, Fesik, S.W. | | Deposit date: | 2024-04-09 | | Release date: | 2024-08-07 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Discovery of a Myeloid Cell Leukemia 1 (Mcl-1) Inhibitor That Demonstrates Potent In Vivo Activities in Mouse Models of Hematological and Solid Tumors.
J.Med.Chem., 67, 2024
|
|
6NW2
 
 | | Structure of human RIPK1 kinase domain in complex with compound 11 | | Descriptor: | (5R)-5-methyl-N-[(3S)-5-methyl-4-oxo-2,3,4,5-tetrahydro-1,5-benzoxazepin-3-yl]-4,5,6,7-tetrahydro-2H-indazole-3-carboxamide, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Fong, R, Lupardus, P.J. | | Deposit date: | 2019-02-05 | | Release date: | 2019-05-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Potent and selective inhibitors of receptor-interacting protein kinase 1 that lack an aromatic back pocket group.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
9GIZ
 
 | | UMG-SP-1, a promiscuous hydrolase | | Descriptor: | GLYCEROL, HEXAETHYLENE GLYCOL, UMG-SP-1 | | Authors: | Tokoli, A, Stojanovski, G, Jodaitis, L, Hollfelder, F, Hyvonen, M. | | Deposit date: | 2024-08-20 | | Release date: | 2025-09-03 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structure of UMG-SP-1 at 1.89 Angstroms resolution
To Be Published
|
|
6ZDK
 
 | | Structure of the catalytic domain of human endo-alpha-mannosidase MANEA in complex with HEPES and hexatungstotellurate(VI) TEW | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 6-tungstotellurate(VI), Glycoprotein endo-alpha-1,2-mannosidase, ... | | Authors: | Sobala, L.F, Fernandes, P.Z, Hakki, Z, Thompson, A.J, Howe, J.D, Hill, M, Zitzmann, N, Davies, S, Stamataki, Z, Butters, T.D, Alonzi, D.S, Williams, S.J, Davies, G.J. | | Deposit date: | 2020-06-14 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of human endo-alpha-1,2-mannosidase (MANEA), an antiviral host-glycosylation target.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5KRE
 
 | | Covalent inhibitor of LYPLAL1 | | Descriptor: | (2~{R})-2-phenylpiperidine-1-carbaldehyde, Lysophospholipase-like protein 1, NITRATE ION | | Authors: | Pandit, J. | | Deposit date: | 2016-07-07 | | Release date: | 2016-07-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of a Selective Covalent Inhibitor of Lysophospholipase-like 1 (LYPLAL1) as a Tool to Evaluate the Role of this Serine Hydrolase in Metabolism.
Acs Chem.Biol., 11, 2016
|
|
