7LRO
 
 | | Crystal structure of BPTF bromodomain in complex with inhibitor HZ-01-105 | | Descriptor: | 1,2-ETHANEDIOL, 5-(azetidin-3-ylamino)-4-chloranyl-2-methyl-pyridazin-3-one, DIMETHYL SULFOXIDE, ... | | Authors: | Chan, A, Schonbrunn, E. | | Deposit date: | 2021-02-17 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
7LPK
 
 | | Crystal structure of BPTF bromodomain in complex with inhibitor HZ-03-112 | | Descriptor: | 1,2-ETHANEDIOL, 4-chloranyl-2-methyl-5-[[(3~{R})-pyrrolidin-3-yl]amino]pyridazin-3-one, Nucleosome-remodeling factor subunit BPTF | | Authors: | Chan, A, Schonbrunn, E. | | Deposit date: | 2021-02-12 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
7TL5
 
 | | Crystal structure of putative hydrolase yjcS from Klebsiella pneumoniae. | | Descriptor: | 1,2-ETHANEDIOL, Lactamase_B domain-containing protein | | Authors: | Chang, C, Endres, M, Wu, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-18 | | Release date: | 2022-02-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
9CTU
 
 | | Cryo-EM structure of SARS-CoV-2 M (short conformation)bound to C1P | | Descriptor: | (2S,3R,4E)-2-(hexadecanoylamino)-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, Membrane protein, POTASSIUM ION, ... | | Authors: | Dolan, K.A, Brohawn, S.G. | | Deposit date: | 2024-07-25 | | Release date: | 2024-11-06 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Direct lipid interactions control SARS-CoV-2 M protein conformational dynamics and virus assembly.
Biorxiv, 2024
|
|
5VFN
 
 | |
5IGK
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with bromosporine (BSP) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, Bromosporine | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, von Delft, F, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-02-28 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Promiscuous targeting of bromodomains by bromosporine identifies BET proteins as master regulators of primary transcription response in leukemia.
Sci Adv, 2, 2016
|
|
4MC3
 
 | | Hedycaryol synthase in complex with Nerolidol | | Descriptor: | (3R,6E)-3,7,11-trimethyldodeca-1,6,10-trien-3-ol, Putative sesquiterpene cyclase | | Authors: | Baer, P, Rabe, P, Cirton, C, Oliveira Mann, C, Kaufmann, N, Groll, M, Dickschat, J. | | Deposit date: | 2013-08-21 | | Release date: | 2014-01-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Hedycaryol synthase in complex with nerolidol reveals terpene cyclase mechanism.
Chembiochem, 15, 2014
|
|
9CLO
 
 | | DosP R97A with c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), MAGNESIUM ION, Oxygen sensor protein DosP, ... | | Authors: | Kumar, P, Kober, D.L. | | Deposit date: | 2024-07-11 | | Release date: | 2024-11-20 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of the multi-domain oxygen sensor DosP: remote control of a c-di-GMP phosphodiesterase by a regulatory PAS domain.
Nat Commun, 15, 2024
|
|
8VXX
 
 | | Mango II bound to 365A-061 | | Descriptor: | 3-[2-({[(6P)-2-fluoro-6-(1H-pyrazol-1-yl)phenyl]methyl}amino)-2-oxoethyl]-2-[(E)-(1-methylquinolin-4(1H)-ylidene)methyl]-1,3-benzothiazol-3-ium, Chains: A,B,C, POTASSIUM ION | | Authors: | Passalacqua, L.F.M, Ferre-D'Amare, A.R, Schneekloth, J.S. | | Deposit date: | 2024-02-06 | | Release date: | 2025-02-19 | | Last modified: | 2025-10-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure-informed design of an ultrabright RNA-activated fluorophore.
Nat.Chem., 17, 2025
|
|
8VY0
 
 | | Mango II bound to 365A-087 | | Descriptor: | 2-[(E)-(1-methylquinolin-4(1H)-ylidene)methyl]-3-[2-oxo-2-({[(2P)-2-(1H-pyrazol-1-yl)phenyl]methyl}amino)ethyl]-1,3-benzothiazol-3-ium, Chains: A,B,C, POTASSIUM ION | | Authors: | Passalacqua, L.F.M, Ferre-D'Amare, A.R, Schneekloth, J.S. | | Deposit date: | 2024-02-06 | | Release date: | 2025-02-19 | | Last modified: | 2025-10-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure-informed design of an ultrabright RNA-activated fluorophore.
Nat.Chem., 17, 2025
|
|
5IDQ
 
 | |
3GPI
 
 | | Structure of putative NAD-dependent epimerase/dehydratase from methylobacillus flagellatus | | Descriptor: | 1,2-ETHANEDIOL, NAD-dependent epimerase/dehydratase | | Authors: | Ramagopal, U.A, Morano, C, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-23 | | Release date: | 2009-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Structure of putative NAD-dependent epimerase/dehydratase from methylobacillus flagellatus
To be published
|
|
1KHI
 
 | | CRYSTAL STRUCTURE OF HEX1 | | Descriptor: | Hex1 | | Authors: | Yuan, P, Swaminathan, K. | | Deposit date: | 2001-11-30 | | Release date: | 2002-11-30 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | A HEX-1 crystal lattice required for Woronin body function in Neurospora crassa
NAT.STRUCT.BIOL., 10, 2003
|
|
8VXZ
 
 | | Mango II bound to 365A-084 | | Descriptor: | 3-[2-(benzylamino)-2-oxoethyl]-2-[(E)-(1-methylquinolin-4(1H)-ylidene)methyl]-1,3-benzothiazol-3-ium, Chains: A,B,C, POTASSIUM ION | | Authors: | Passalacqua, L.F.M, Ferre-D'Amare, A.R, Schneekloth, J.S. | | Deposit date: | 2024-02-06 | | Release date: | 2025-02-19 | | Last modified: | 2025-10-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-informed design of an ultrabright RNA-activated fluorophore.
Nat.Chem., 17, 2025
|
|
8VY1
 
 | | Mango II bound to 365A-088 | | Descriptor: | 3-(2-{[(2-fluorophenyl)methyl]amino}-2-oxoethyl)-2-[(E)-(1-methylquinolin-4(1H)-ylidene)methyl]-1,3-benzothiazol-3-ium, Chains: A,B,C, POTASSIUM ION | | Authors: | Passalacqua, L.F.M, Ferre-D'Amare, A.R, Schneekloth, J.S. | | Deposit date: | 2024-02-06 | | Release date: | 2025-02-19 | | Last modified: | 2025-10-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure-informed design of an ultrabright RNA-activated fluorophore.
Nat.Chem., 17, 2025
|
|
1SIZ
 
 | |
3GSJ
 
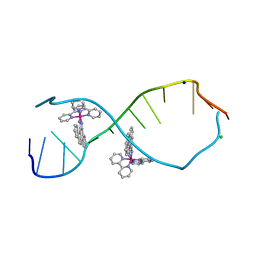 | | A Bulky Rhodium Complex Bound to an Adenosine-Adenosine DNA Mismatch | | Descriptor: | 5'-D(*CP*GP*GP*AP*AP*AP*TP*TP*AP*CP*CP*G)-3', CHLORIDE ION, SODIUM ION, ... | | Authors: | Zeglis, B.M, Pierre, V.C, Kaiser, J.T, Barton, J.K. | | Deposit date: | 2009-03-27 | | Release date: | 2009-05-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A bulky rhodium complex bound to an adenosine-adenosine DNA mismatch: general architecture of the metalloinsertion binding mode
Biochemistry, 48, 2009
|
|
5VEV
 
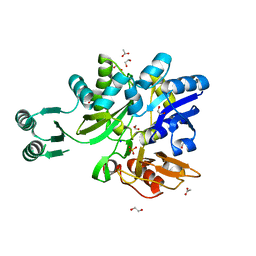 | |
5IS5
 
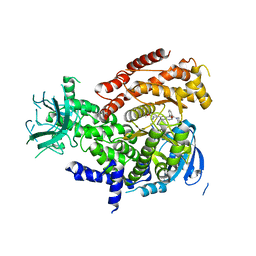 | |
9CMF
 
 | | Substrate bound DosP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), MAGNESIUM ION, OXYGEN MOLECULE, ... | | Authors: | Kumar, P, Kober, D.L. | | Deposit date: | 2024-07-15 | | Release date: | 2024-11-20 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Structures of the multi-domain oxygen sensor DosP: remote control of a c-di-GMP phosphodiesterase by a regulatory PAS domain.
Nat Commun, 15, 2024
|
|
2XP2
 
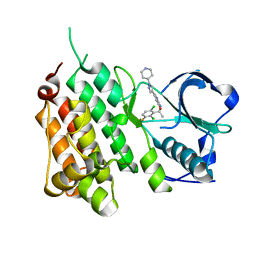 | | Structure of the Human Anaplastic Lymphoma Kinase in Complex with Crizotinib (PF-02341066) | | Descriptor: | 3-[(1R)-1-(2,6-dichloro-3-fluorophenyl)ethoxy]-5-(1-piperidin-4-yl-1H-pyrazol-4-yl)pyridin-2-amine, TYROSINE-PROTEIN KINASE RECEPTOR | | Authors: | McTigue, M, Deng, Y, Liu, W, Brooun, A, Timofeevski, S, Marrone, T, Cui, J.J. | | Deposit date: | 2010-08-24 | | Release date: | 2010-09-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure Based Drug Design of Crizotinib (Pf-02341066), a Potent and Selective Dual Inhibitor of Mesenchymal-Epithelial Transition Factor (C-met) Kinase and Anaplastic Lymphoma Kinase (Alk).
J.Med.Chem, 54, 2011
|
|
5VW8
 
 | | NADP+ soak of Y316S mutant of corn root ferredoxin:NADP+ reductase in alternate space group | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin--NADP reductase, MAGNESIUM ION, ... | | Authors: | Kean, K.M, Carpenter, R.A, Hall, A.R, Karplus, P.A. | | Deposit date: | 2017-05-21 | | Release date: | 2017-08-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.808 Å) | | Cite: | High-resolution studies of hydride transfer in the ferredoxin:NADP(+) reductase superfamily.
FEBS J., 284, 2017
|
|
4DN5
 
 | | Crystal Structure of NF-kB-inducing Kinase (NIK) | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Min, X, Liu, J, Sudom, A, Walker, N.P, Wang, Z. | | Deposit date: | 2012-02-08 | | Release date: | 2012-06-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Nuclear Factor Kappa B-inducing kinase domain reveals a constitutively active conformation
J.Biol.Chem., 287, 2012
|
|
2Y54
 
 | | Fragment growing induces conformational changes in acetylcholine- binding protein: A structural and thermodynamic analysis - (Fragment 1) | | Descriptor: | CHLORIDE ION, SOLUBLE ACETYLCHOLINE RECEPTOR, SULFATE ION, ... | | Authors: | Rucktooa, P, Edink, E, deEsch, I.J.P, Sixma, T.K. | | Deposit date: | 2011-01-12 | | Release date: | 2011-06-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Fragment Growing Induces Conformational Changes in Acetylcholine-Binding Protein: A Structural and Thermodynamic Analysis.
J.Am.Chem.Soc., 133, 2011
|
|
7ATL
 
 | | EstCE1, a hydrolase with promiscuous acyltransferase activity | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Esterase, ... | | Authors: | Palm, G.J, Lammers, M, Berndt, L. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.478 Å) | | Cite: | Discovery and Design of Family VIII Carboxylesterases as Highly Efficient Acyltransferases.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
