1NIB
 
 | | THE STRUCTURE OF CU-NITRITE REDUCTASE FROM ACHROMOBACTER CYCLOCLASTES AT FIVE PH VALUES, WITH NITRITE BOUND AND WITH TYPE II CU DEPLETED | | Descriptor: | COPPER (II) ION, NITRITE REDUCTASE | | Authors: | Adman, E.T, Godden, J.W, Turley, S. | | Deposit date: | 1995-07-03 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structure of copper-nitrite reductase from Achromobacter cycloclastes at five pH values, with NO2- bound and with type II copper depleted.
J.Biol.Chem., 270, 1995
|
|
1NIE
 
 | | THE STRUCTURE OF CU-NITRITE REDUCTASE FROM ACHROMOBACTER CYCLOCLASTES AT FIVE PH VALUES, WITH NITRITE BOUND AND WITH TYPE II CU DEPLETED | | Descriptor: | COPPER (II) ION, NITRITE REDUCTASE | | Authors: | Adman, E.T, Godden, J.W, Turley, S. | | Deposit date: | 1995-07-03 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of copper-nitrite reductase from Achromobacter cycloclastes at five pH values, with NO2- bound and with type II copper depleted.
J.Biol.Chem., 270, 1995
|
|
1OF0
 
 | | CRYSTAL STRUCTURE OF BACILLUS SUBTILIS COTA AFTER 1H SOAKING WITH ABTS | | Descriptor: | 3-ETHYL-2-[(2Z)-2-(3-ETHYL-6-SULFO-1,3-BENZOTHIAZOL-2(3H)-YLIDENE)HYDRAZINO]-6-SULFO-3H-1,3-BENZOTHIAZOL-1-IUM, COPPER (I) ION, CU-O LINKAGE, ... | | Authors: | Enguita, F.J, Marcal, D, Grenha, R, Martins, L.O, Henriques, A.O, Carrondo, M.A. | | Deposit date: | 2003-04-03 | | Release date: | 2004-05-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural Characterization of a Bacterial Laccase Reaction Cycle
To be Published
|
|
1NPJ
 
 | | Crystal structure of H145A mutant of nitrite reductase from Alcaligenes faecalis | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase | | Authors: | Wijma, H.J, Boulanger, M.J, Molon, A, Fittipaldi, M, Huber, M, Murphy, M.E, Verbeet, M.P, Canters, G.W. | | Deposit date: | 2003-01-18 | | Release date: | 2003-04-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Reconstitution of the type-1 active site of the H145G/A variants of nitrite reductase by ligand insertion
Biochemistry, 42, 2003
|
|
1N68
 
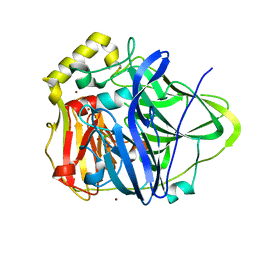 | | Copper bound to the Multicopper Oxidase CueO | | Descriptor: | Blue copper oxidase cueO, COPPER (II) ION, CU-CL-CU LINKAGE | | Authors: | Roberts, S.A, Wildner, G.F, Grass, G, Weichsel, A, Ambrus, A, Rensing, C, Montfort, W.R. | | Deposit date: | 2002-11-08 | | Release date: | 2003-06-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Labile Regulatory Copper Ion Lies Near the T1 Copper Site in the Multicopper Oxidase CueO.
J.Biol.Chem., 278, 2003
|
|
1N70
 
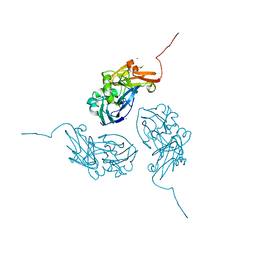 | |
2XX0
 
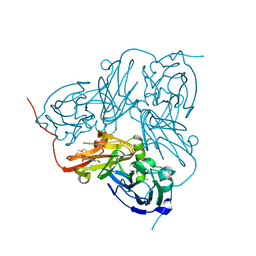 | | STRUCTURE OF THE N90S-H254F MUTANT OF NITRITE REDUCTASE FROM ALCALIGENES XYLOSOXIDANS | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, ... | | Authors: | Antonyuk, S.V, Leferink, N.G.H, Han, C, Heyes, D.J, Rigby, S.E.J, Hough, M.A, Eady, R.R, Scrutton, N.S, Hasnain, S.S. | | Deposit date: | 2010-11-07 | | Release date: | 2011-05-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Proton-Coupled Electron Transfer in the Catalytic Cycle of Alcaligenes Xylosoxidans Copper-Dependent Nitrite Reductase.
Biochemistry, 50, 2011
|
|
2XVB
 
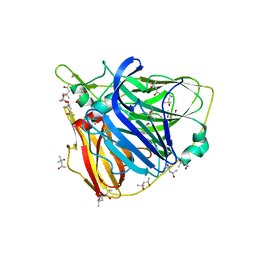 | | Crystal structure of Laccase from Thermus thermophilus HB27 complexed with Hg, crystal of the apoenzyme soaked for 5 min. in 5 mM HgCl2 at 278 K. | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, LACCASE, ... | | Authors: | Serrano-Posada, H, Valderrama, B, Rudino-Pinera, E. | | Deposit date: | 2010-10-25 | | Release date: | 2011-10-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-Ray-Induced Catalytic Active-Site Reduction of a Multicopper Oxidase: Structural Insights Into the Proton-Relay Mechanism and O2-Reduction States.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2YAF
 
 | | X-ray induced reduction of laccase from Thermus thermophilus HB27 (12. 5-25.0 percent dose) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, ... | | Authors: | Serrano-Posada, H, Rudino-Pinera, E. | | Deposit date: | 2011-02-22 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-Ray-Induced Catalytic Active-Site Reduction of a Multicopper Oxidase: Structural Insights Into the Proton-Relay Mechanism and O2-Reduction States.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2YAP
 
 | | X-ray induced reduction of laccase from Thermus thermophilus HB27 (62. 5-75.0 percent dose) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, ... | | Authors: | Serrano-Posada, H, Rudino-Pinera, E. | | Deposit date: | 2011-02-23 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-Ray-Induced Catalytic Active-Site Reduction of a Multicopper Oxidase: Structural Insights Into the Proton-Relay Mechanism and O2-Reduction States.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2YQB
 
 | | Structure of P93A variant of three-domain heme-Cu nitrite reductase from Ralstonia pickettii at 1.4 A resolution | | Descriptor: | COPPER (II) ION, COPPER-CONTAINING NITRITE REDUCTASE, HEME C | | Authors: | Antonyuk, S.V, Han, C, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2012-11-07 | | Release date: | 2013-04-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structures of protein-protein complexes involved in electron transfer.
Nature, 496, 2013
|
|
2Y1A
 
 | |
1OE2
 
 | | Atomic Resolution Structure of D92E Mutant of Alcaligenes xylosoxidans Nitrite Reductase | | Descriptor: | COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, TETRAETHYLENE GLYCOL | | Authors: | Ellis, M.J, Dodd, F.E, Sawers, G, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2003-03-18 | | Release date: | 2003-04-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Atomic Resolution Structures of Native Copper Nitrite Reductase from Alcaligenes Xylosoxidans and the Active Site Mutant Asp92Glu
J.Mol.Biol., 328, 2003
|
|
2YAH
 
 | | X-ray induced reduction of laccase from Thermus thermophilus HB27 (25. 0-37.5 percent dose) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, ... | | Authors: | Serrano-Posada, H, Rudino-Pinera, E. | | Deposit date: | 2011-02-22 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-Ray-Induced Catalytic Active-Site Reduction of a Multicopper Oxidase: Structural Insights Into the Proton-Relay Mechanism and O2-Reduction States.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2YAE
 
 | | X-ray induced reduction of laccase from Thermus thermophilus HB27(0.0- 12.5 percent dose) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, ... | | Authors: | Serrano-Posada, H, Rudino-Pinera, E. | | Deposit date: | 2011-02-21 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-Ray-Induced Catalytic Active-Site Reduction of a Multicopper Oxidase: Structural Insights Into the Proton-Relay Mechanism and O2-Reduction States.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2YAO
 
 | | X-ray induced reduction of laccase from Thermus thermophilus HB27 (50. 0-62.5 percent dose) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, ... | | Authors: | Serrano-Posada, H, Rudino-Pinera, E. | | Deposit date: | 2011-02-23 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-Ray-Induced Catalytic Active-Site Reduction of a Multicopper Oxidase: Structural Insights Into the Proton-Relay Mechanism and O2-Reduction States.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2YAR
 
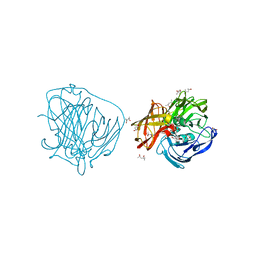 | | X-ray induced reduction of laccase from Thermus thermophilus HB27 (87. 5-100.0 percent dose) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, ... | | Authors: | Serrano-Posada, H, Rudino-Pinera, E. | | Deposit date: | 2011-02-24 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-Ray-Induced Catalytic Active-Site Reduction of a Multicopper Oxidase: Structural Insights Into the Proton-Relay Mechanism and O2-Reduction States.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2XXF
 
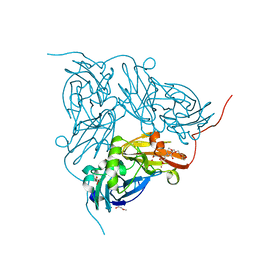 | | Cu metallated H254F mutant of nitrite reductase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, COPPER (II) ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Hough, M.A, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2010-11-10 | | Release date: | 2011-05-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Proton-Coupled Electron Transfer in the Catalytic Cycle of Alcaligenes Xylosoxidans Copper-Dependent Nitrite Reductase.
Biochemistry, 50, 2011
|
|
1NTD
 
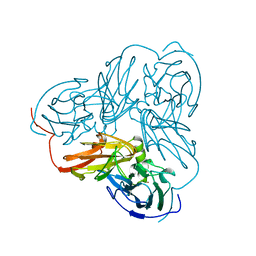 | | STRUCTURE OF ALCALIGENES FAECALIS NITRITE REDUCTASE MUTANT M150E THAT CONTAINS ZINC | | Descriptor: | COPPER (II) ION, NITRITE REDUCTASE | | Authors: | Murphy, M.E.P, Adman, E.T, Turley, S. | | Deposit date: | 1995-07-03 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Alcaligenes faecalis nitrite reductase and a copper site mutant, M150E, that contains zinc.
Biochemistry, 34, 1995
|
|
2XWZ
 
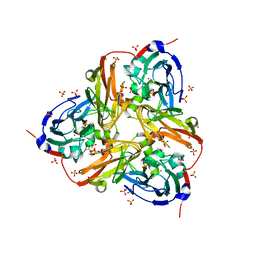 | | STRUCTURE OF THE RECOMBINANT NATIVE NITRITE REDUCTASE FROM ALCALIGENES XYLOSOXIDANS complexed with nitrite | | Descriptor: | ACETATE ION, COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, ... | | Authors: | Antonyuk, S.V, Leferink, N.G.H, Han, C, Heyes, D.J, Rigby, S.E.J, Hough, M.A, Eady, R.R, Scrutton, N.S, Hasnain, S.S. | | Deposit date: | 2010-11-06 | | Release date: | 2011-05-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Proton-Coupled Electron Transfer in the Catalytic Cycle of Alcaligenes Xylosoxidans Copper-Dependent Nitrite Reductase.
Biochemistry, 50, 2011
|
|
2ZON
 
 | | Crystal structure of electron transfer complex of nitrite reductase with cytochrome c | | Descriptor: | COPPER (II) ION, Dissimilatory copper-containing nitrite reductase, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Nojiri, M, Koteishi, H, Yamaguchi, K, Suzuki, S. | | Deposit date: | 2008-05-27 | | Release date: | 2009-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of inter-protein electron transfer for nitrite reduction in denitrification
Nature, 462, 2009
|
|
2YXV
 
 | | The deletion mutant of Multicopper Oxidase CueO | | Descriptor: | Blue copper oxidase cueO, COPPER (II) ION, CU-O-CU LINKAGE, ... | | Authors: | Higuchi, Y, Komori, H. | | Deposit date: | 2007-04-27 | | Release date: | 2008-01-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structure and function of the engineered multicopper oxidase CueO from Escherichia coli--deletion of the methionine-rich helical region covering the substrate-binding site
J.Mol.Biol., 373, 2007
|
|
1PF3
 
 | | Crystal Structure of the M441L mutant of the multicopper oxidase CueO | | Descriptor: | Blue copper oxidase cueO, COPPER (II) ION, CU-CL-CU LINKAGE | | Authors: | Roberts, S.A, Wildner, G.F, Grass, G, Weichsel, A, Ambrus, A, Rensing, C, Montfort, W.R. | | Deposit date: | 2003-05-23 | | Release date: | 2003-06-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Labile Regulatory Copper Ion Lies Near the T1 Copper Site in the Multicopper Oxidase CueO.
J.Biol.Chem., 278, 2003
|
|
3ABG
 
 | | X-ray Crystal Analysis of Bilirubin Oxidase from Myrothecium verrucaria at 2.3 angstrom Resolution using a Twin Crystal | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Bilirubin oxidase, COPPER (II) ION, ... | | Authors: | Mizutani, K, Toyoda, M, Sagara, K, Takahashi, N, Sato, A, Kamitaka, Y, Tsujimura, S, Nakanishi, Y, Sugiura, T, Yamaguchi, S, Kano, K, Mikami, B. | | Deposit date: | 2009-12-10 | | Release date: | 2010-08-18 | | Last modified: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray analysis of bilirubin oxidase from Myrothecium verrucaria at 2.3 A resolution using a twinned crystal
Acta Crystallogr.,Sect.F, 66, 2010
|
|
1NPN
 
 | | Crystal structure of a copper reconstituted H145A mutant of nitrite reductase from Alcaligenes faecalis | | Descriptor: | CHLORIDE ION, COPPER (II) ION, Copper-containing nitrite reductase | | Authors: | Wijma, H.J, Boulanger, M.J, Molon, A, Fittipaldi, M, Huber, M, Murphy, M.E, Verbeet, M.P, Canters, G.W. | | Deposit date: | 2003-01-18 | | Release date: | 2003-04-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Reconstitution of the type-1 active site of the 145G/A variants of Nitrite Reductase by ligand insertion
Biochemistry, 42, 2003
|
|
