6HQ7
 
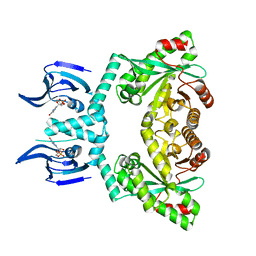 | | Structure of EAL Enzyme Bd1971 - cGMP bound form | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, EAL Enzyme Bd1971, MAGNESIUM ION | | Authors: | Lovering, A.L, Cadby, I.T. | | Deposit date: | 2018-09-24 | | Release date: | 2019-07-31 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Nucleotide signaling pathway convergence in a cAMP-sensing bacterial c-di-GMP phosphodiesterase.
Embo J., 38, 2019
|
|
9R2M
 
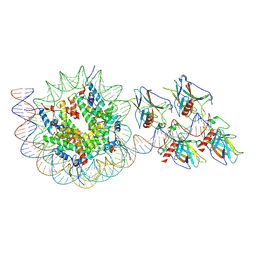 | | p53 bound to nucleosome at position SHL+5.9 (non-crosslinked sample, composite map) | | Descriptor: | Cellular tumor antigen p53, DNA-for, DNA-rev, ... | | Authors: | Chakraborty, D, Kater, L, Kempf, G, Cavadini, S, Thoma, N.H. | | Deposit date: | 2025-04-30 | | Release date: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | p53 bound to nucleosome at position SHL+5.9 (non-crosslinked sample, composite map)
To Be Published
|
|
6F4D
 
 | |
7BX2
 
 | |
7S3F
 
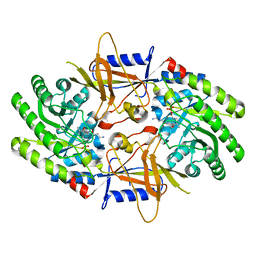 | | Structure of cofactor pyridoxal 5-phosphate bound human ornithine decarboxylase in complex with its inhibitor 1-amino-oxy-3-aminopropane | | Descriptor: | 3-AMINOOXY-1-AMINOPROPANE, Ornithine decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Zhou, X.E, Suino-Powell, K, Schultz, C.R, Aleiwi, B, Brunzelle, J.S, Lamp, J, Vega, I.E, Ellsworth, E, Bachmann, A.S, Melcher, K. | | Deposit date: | 2021-09-06 | | Release date: | 2021-12-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural basis of binding and inhibition of ornithine decarboxylase by 1-amino-oxy-3-aminopropane.
Biochem.J., 478, 2021
|
|
7S3G
 
 | | Structure of cofactor pyridoxal 5-phosphate bound human ornithine decarboxylase in complex with citrate at the catalytic center | | Descriptor: | CITRIC ACID, Ornithine decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Zhou, X.E, Suino-Powell, K, Schultz, C.R, Aleiwi, B, Brunzelle, J.S, Lamp, J, Vega, I.E, Ellsworth, E, Bachmann, A.S, Melcher, K. | | Deposit date: | 2021-09-06 | | Release date: | 2021-12-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural basis of binding and inhibition of ornithine decarboxylase by 1-amino-oxy-3-aminopropane.
Biochem.J., 478, 2021
|
|
6G74
 
 | |
9GGR
 
 | | Structure of the HrpA-bound E. coli disome, Class II | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 50S ribosomal protein L11, ... | | Authors: | Esser, H.F, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2024-08-14 | | Release date: | 2025-02-12 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The RNA helicase HrpA rescues collided ribosomes in E. coli.
Mol.Cell, 85, 2025
|
|
2L52
 
 | | Solution structure of the small archaeal modifier protein 1 (SAMP1) from Methanosarcina acetivorans | | Descriptor: | METHANOSARCINA ACETIVORANS SAMP1 HOMOLOG | | Authors: | Damberger, F.F, Ranjan, N, Sutter, M, Allain, F.H.-T, Weber-Ban, E. | | Deposit date: | 2010-10-24 | | Release date: | 2011-02-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure and activation mechanism of ubiquitin-like small archaeal modifier proteins.
J.Mol.Biol., 405, 2011
|
|
9HPC
 
 | |
9HPE
 
 | |
5IO2
 
 | | Xanthomonas campestris Peroxiredoxin Q - C48S mutant | | Descriptor: | Bacterioferritin comigratory protein, PHOSPHATE ION, SODIUM ION | | Authors: | Perkins, A, Parsonage, D, Nelson, K.J, Poole, L.B, Karplus, A. | | Deposit date: | 2016-03-08 | | Release date: | 2016-09-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Peroxiredoxin Catalysis at Atomic Resolution.
Structure, 24, 2016
|
|
6GIV
 
 | | Structure of GluA2-N775S ligand-binding domain (S1S2J) in complex with glutamate and Rubidium Bromide at 1.75 A resolution | | Descriptor: | BROMIDE ION, GLUTAMIC ACID, GLYCEROL, ... | | Authors: | Venskutonyte, R, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2018-05-15 | | Release date: | 2019-05-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Nanoscale Mobility of the Apo State and TARP Stoichiometry Dictate the Gating Behavior of Alternatively Spliced AMPA Receptors.
Neuron, 102, 2019
|
|
8UNJ
 
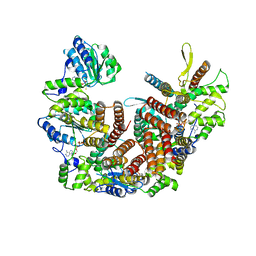 | | Atomic model of the human CTF18-RFC alone in the apo state (State 1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chromosome transmission fidelity protein 18 homolog, MAGNESIUM ION, ... | | Authors: | Wang, F, He, Q, Li, H. | | Deposit date: | 2023-10-19 | | Release date: | 2024-05-08 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Cryo-EM reveals a nearly complete PCNA loading process and unique features of the human alternative clamp loader CTF18-RFC.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6U5S
 
 | |
6UCB
 
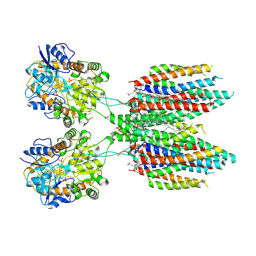 | | GluA2 in complex with its auxiliary subunit CNIH3 - with antagonist ZK200775, LBD, TMD, CNIH3, and lipids | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHOLESTEROL, Glutamate receptor 2, ... | | Authors: | Nakagawa, T. | | Deposit date: | 2019-09-15 | | Release date: | 2019-12-04 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structures of the AMPA receptor in complex with its auxiliary subunit cornichon.
Science, 366, 2019
|
|
8FWA
 
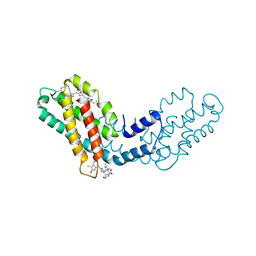 | | Phycocyanin structure from a modular droplet injector for serial femtosecond crystallography | | Descriptor: | C-phycocyanin alpha chain, C-phycocyanin beta chain, PHYCOCYANOBILIN, ... | | Authors: | Botha, S, Doppler, D.L, Sonker, M, Egatz-Gomez, A, Grieco, A, Zaare, S, Jernigan, R, Meza-Aguilar, J.D, Rabbani, M.T, Manna, A, Alvarez, R, Karpos, K, Cruz Villarreal, J, Nelson, G, Ketawala, G.K, Pey, A.L, Ruiz-Fresneda, M.A, Pacheco-Garcia, J.L, Nazari, R, Sierra, R, Hunter, M.S, Batyuk, A, Kupitz, C.J, Sublett, R.E, Lisova, S, Mariani, V, Boutet, S, Fromme, R, Grant, T.D, Fromme, P, Kirian, R.A, Martin-Garcia, J.M, Ros, A. | | Deposit date: | 2023-01-20 | | Release date: | 2023-06-28 | | Last modified: | 2023-07-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Modular droplet injector for sample conservation providing new structural insight for the conformational heterogeneity in the disease-associated NQO1 enzyme.
Lab Chip, 23, 2023
|
|
6UD8
 
 | |
9HPD
 
 | |
9QFH
 
 | |
9HPF
 
 | |
6RRL
 
 | |
6RSM
 
 | | Solution NMR structure of the peptide 12530 from medicinal leech Hirudo medicinalis in dodecylphosphocholine micelles | | Descriptor: | peptide 12530 | | Authors: | Talyzina, I.A, Nadezhdin, K.D, Grafskaia, E.N, Arseniev, A.S, Lazarev, V.N. | | Deposit date: | 2019-05-21 | | Release date: | 2019-07-24 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Medicinal leech antimicrobial peptides lacking toxicity represent a promising alternative strategy to combat antibiotic-resistant pathogens.
Eur.J.Med.Chem., 180, 2019
|
|
5IOX
 
 | | Xanthomonas campestris Peroxiredoxin Q - Structure LUss | | Descriptor: | Bacterioferritin comigratory protein | | Authors: | Perkins, A, Parsonage, D, Nelson, K.J, Poole, L.B, Karplus, A. | | Deposit date: | 2016-03-09 | | Release date: | 2016-09-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Peroxiredoxin Catalysis at Atomic Resolution.
Structure, 24, 2016
|
|
5IM9
 
 | | Xanthomonas campestris Peroxiredoxin Q - Structure F1 | | Descriptor: | Bacterioferritin comigratory protein, SODIUM ION | | Authors: | Perkins, A, Parsonage, D, Nelson, K.J, Poole, L.B, Karplus, A. | | Deposit date: | 2016-03-05 | | Release date: | 2016-09-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Peroxiredoxin Catalysis at Atomic Resolution.
Structure, 24, 2016
|
|
