6GNE
 
 | | Catalytic domain of Starch Synthase IV from Arabidopsis thaliana bound to ADP and acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, Probable starch synthase 4, ... | | Authors: | Cuesta-Seijo, J.A, Ruzanski, C, Krucewicz, K, Striebeck, A, Palcic, M.M. | | Deposit date: | 2018-05-30 | | Release date: | 2018-07-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal Structures of theCatalyticDomain ofArabidopsis thalianaStarch Synthase IV, of Granule Bound Starch Synthase From CLg1 and of Granule Bound Starch Synthase I ofCyanophora paradoxaIllustrate Substrate Recognition in Starch Synthases.
Front Plant Sci, 9, 2018
|
|
6GOL
 
 | |
1ZZL
 
 | | Crystal structure of P38 with triazolopyridine | | Descriptor: | 6-[4-(4-FLUOROPHENYL)-1,3-OXAZOL-5-YL]-3-ISOPROPYL[1,2,4]TRIAZOLO[4,3-A]PYRIDINE, Mitogen-activated protein kinase 14 | | Authors: | McClure, K.F, Han, S. | | Deposit date: | 2005-06-14 | | Release date: | 2005-09-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Theoretical and Experimental Design of Atypical Kinase Inhibitors: Application to p38 MAP Kinase.
J.Med.Chem., 48, 2005
|
|
3HV6
 
 | | Human p38 MAP Kinase in Complex with RL39 | | Descriptor: | 1-[3-tert-butyl-1-(4-methylphenyl)-1H-pyrazol-5-yl]-3-[4-(2-morpholin-4-ylethoxy)phenyl]urea, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Gruetter, C, Simard, J.R, Getlik, M, Rauh, D. | | Deposit date: | 2009-06-15 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Displacement assay for the detection of stabilizers of inactive kinase conformations.
J.Med.Chem., 53, 2010
|
|
6HYG
 
 | | Heteromeric tandem IgG4/IgG1 Fc | | Descriptor: | IgHG1 and IgHG4 hybrid, ZINC ION | | Authors: | Casaletto, J.B, Geddie, M.L, Abu-Yousif, A.O, Masson, K, Fulgham, A, Boudot, A, Maiwald, T, Kearns, J.D, Kohli, N, Su, S, Razlog, M, Raue, A, Kalra, A, Hakansson, M, Logan, D.T, Welin, M, Chattopadhyay, S, Harms, B.D, Nielsen, U.B, Schoeberl, B, Lugovskoy, A.A, MacBeath, G. | | Deposit date: | 2018-10-21 | | Release date: | 2019-03-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | MM-131, a bispecific anti-Met/EpCAM mAb, inhibits HGF-dependent and HGF-independent Met signaling through concurrent binding to EpCAM.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6GIV
 
 | | Structure of GluA2-N775S ligand-binding domain (S1S2J) in complex with glutamate and Rubidium Bromide at 1.75 A resolution | | Descriptor: | BROMIDE ION, GLUTAMIC ACID, GLYCEROL, ... | | Authors: | Venskutonyte, R, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2018-05-15 | | Release date: | 2019-05-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Nanoscale Mobility of the Apo State and TARP Stoichiometry Dictate the Gating Behavior of Alternatively Spliced AMPA Receptors.
Neuron, 102, 2019
|
|
6H0H
 
 | | The ABC transporter associated binding protein from B. animalis subsp. lactis Bl-04 in complex with beta-1,6-galactobiose | | Descriptor: | DI(HYDROXYETHYL)ETHER, Probable solute binding protein of ABC transporter system for sugars, TETRAETHYLENE GLYCOL, ... | | Authors: | Fredslund, F, Lo Leggio, L. | | Deposit date: | 2018-07-09 | | Release date: | 2019-06-19 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Substrate preference of an ABC importer corresponds to selective growth on beta-(1,6)-galactosides inBifidobacterium animalissubsp.lactis.
J.Biol.Chem., 294, 2019
|
|
6I93
 
 | |
6IAO
 
 | | Structure of Cytochrome P450 BM3 M11 mutant in complex with DTT at resolution 2.16A | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Bifunctional cytochrome P450/NADPH--P450 reductase, CHLORIDE ION, ... | | Authors: | Mirza, O, Rafiq, M, Frydenvang, K. | | Deposit date: | 2018-11-27 | | Release date: | 2019-06-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural analysis of Cytochrome P450 BM3 mutant M11 in complex with dithiothreitol.
Plos One, 14, 2019
|
|
1YO5
 
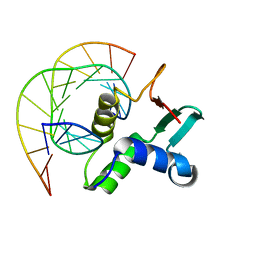 | | Analysis of the 2.0A crystal structure of the protein-DNA complex of human PDEF Ets domain bound to the prostate specific antigen regulatory site | | Descriptor: | Enhancer site of Prostate Specific Antigen Promoter Region, SAM pointed domain containing ets transcription factor | | Authors: | Wang, Y, Feng, L, Said, M, Balderman, S, Fayazi, Z, Liu, Y, Ghosh, D, Gulick, A.M. | | Deposit date: | 2005-01-26 | | Release date: | 2005-05-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Analysis of the 2.0 A Crystal Structure of the Protein-DNA Complex of the Human PDEF Ets Domain Bound to the Prostate Specific Antigen Regulatory Site
Biochemistry, 44, 2005
|
|
6I74
 
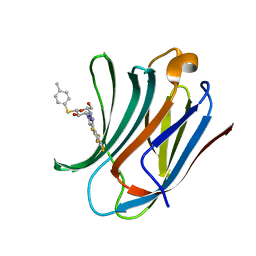 | | Galectin-3C in complex with substituted polyfluoroaryl monothiogalactoside derivative 1 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-(4-methylphenyl)sulfanyl-4-[4-[2,3,4,5,6-pentakis(fluoranyl)phenyl]-1,2,3-triazol-1-yl]oxane-3,5-diol, Galectin-3 | | Authors: | Kumar, R, Peterson, K, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2018-11-15 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.959 Å) | | Cite: | Substituted polyfluoroaryl interactions with an arginine side chain in galectin-3 are governed by steric-, desolvation and electronic conjugation effects.
Org. Biomol. Chem., 17, 2019
|
|
2AAG
 
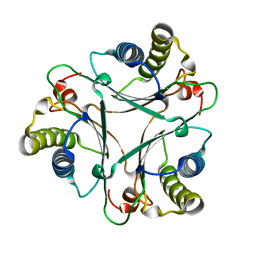 | | Crystal Structures of the Wild-type, Mutant-P1A and Inactivated Malonate Semialdehyde Decarboxylase: A Structural Basis for the Decarboxylase and Hydratase Activities | | Descriptor: | Malonate Semialdehyde Decarboxylase | | Authors: | Almrud, J.J, Poelarends, G.J, Johnson Jr, W.H, Serrano, H, Hackert, M.L, Whitman, C.P. | | Deposit date: | 2005-07-13 | | Release date: | 2005-11-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structures of the Wild-Type, P1A Mutant, and Inactivated Malonate Semialdehyde Decarboxylase: A Structural Basis for the Decarboxylase and Hydratase Activities
Biochemistry, 44, 2005
|
|
6I76
 
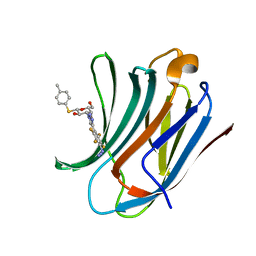 | | Galectin-3C in complex with substituted polyfluoroaryl monothiogalactoside derivative-3 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R},6~{S})-4-[4-[4-azido-2,3,5,6-tetrakis(fluoranyl)phenyl]-1,2,3-triazol-1-yl]-2-(hydroxymethyl)-6-(4-methylphenyl)sulfanyl-oxane-3,5-diol, Galectin-3 | | Authors: | Kumar, R, Peterson, K, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2018-11-15 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Substituted polyfluoroaryl interactions with an arginine side chain in galectin-3 are governed by steric-, desolvation and electronic conjugation effects.
Org. Biomol. Chem., 17, 2019
|
|
6HAQ
 
 | | AFGH61B WILD-TYPE COPPER LOADED | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, COPPER (II) ION, ... | | Authors: | Lo Leggio, L, Poulsen, J.C.N, Weihe, C.D. | | Deposit date: | 2018-08-08 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Structure of a lytic polysaccharide monooxygenase from Aspergillus fumigatus and an engineered thermostable variant.
Carbohydr. Res., 469, 2018
|
|
6I8C
 
 | | Crystal structure of the murine beta-2-microglobulin. | | Descriptor: | Beta-2-microglobulin, TRIETHYLENE GLYCOL | | Authors: | Achour, A, Sandalova, T, Ricagno, S, Sun, R. | | Deposit date: | 2018-11-20 | | Release date: | 2019-10-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Biochemical and biophysical comparison of human and mouse beta-2 microglobulin reveals the molecular determinants of low amyloid propensity.
Febs J., 287, 2020
|
|
6I9X
 
 | | urate oxidase under 35 bar of argon | | Descriptor: | 8-AZAXANTHINE, ARGON, SODIUM ION, ... | | Authors: | Prange, T, Colloc'h, N. | | Deposit date: | 2018-11-26 | | Release date: | 2019-12-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Comparative study of the effects of high hydrostatic pressure per se and high argon pressure on urate oxidase ligand stabilization
Acta Cryst. D, 78, 2022
|
|
6I77
 
 | | Galectin-3C in complex with substituted polyfluoroaryl monothiogalactoside derivative-4 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R},6~{S})-4-[4-[4-azanyl-2,3,5,6-tetrakis(fluoranyl)phenyl]-1,2,3-triazol-1-yl]-2-(hydroxymethyl)-6-(4-methylphenyl)sulfanyl-oxane-3,5-diol, Galectin-3 | | Authors: | Kumar, R, Peterson, K, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2018-11-15 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.219 Å) | | Cite: | Substituted polyfluoroaryl interactions with an arginine side chain in galectin-3 are governed by steric-, desolvation and electronic conjugation effects.
Org. Biomol. Chem., 17, 2019
|
|
2OZQ
 
 | | Crystal Structure of apo-MUP | | Descriptor: | CADMIUM ION, Novel member of the major urinary protein (Mup) gene family, SODIUM ION | | Authors: | Dennis, C.A, Homans, S.W, Phillips, S.E.V, Syme, N.R. | | Deposit date: | 2007-02-27 | | Release date: | 2008-01-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Origin of heat capacity changes in a "nonclassical" hydrophobic interaction.
Chembiochem, 8, 2007
|
|
2NPQ
 
 | |
2PKD
 
 | | Crystal structure of CD84: Insite into SLAM family function | | Descriptor: | CHLORIDE ION, SLAM family member 5 | | Authors: | Yan, Q, Malashkevich, V.N, Fedorov, A, Cao, E, Lary, J.W, Cole, J.L, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2007-04-17 | | Release date: | 2007-06-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.043 Å) | | Cite: | Structure of CD84 provides insight into SLAM family function.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2B13
 
 | | Truncated S. aureus LytM, P41 crystal form | | Descriptor: | Glycyl-glycine endopeptidase lytM, L(+)-TARTARIC ACID, ZINC ION | | Authors: | Firczuk, M, Mucha, A, Bochtler, M. | | Deposit date: | 2005-09-15 | | Release date: | 2006-01-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of active LytM.
J.Mol.Biol., 354, 2005
|
|
8OVN
 
 | | X-ray structure of the SF-iGluSnFR-S72A | | Descriptor: | CITRIC ACID, Putative periplasmic binding transport protein,Green fluorescent protein | | Authors: | Tarnawski, M, Hellweg, L, Bergner, A, Hiblot, J, Leippe, P, Johnsson, K. | | Deposit date: | 2023-04-26 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | X-ray structure of the SF-iGluSnFR-S72A
To Be Published
|
|
8OVO
 
 | | X-ray structure of the SF-iGluSnFR-S72A in complex with L-aspartate | | Descriptor: | ASPARTIC ACID, Putative periplasmic binding transport protein,Green fluorescent protein | | Authors: | Tarnawski, M, Hellweg, L, Bergner, A, Hiblot, J, Leippe, P, Johnsson, K. | | Deposit date: | 2023-04-26 | | Release date: | 2023-05-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray structure of the SF-iGluSnFR-S72A in complex with L-aspartate
To Be Published
|
|
8OVP
 
 | | X-ray structure of the iAspSnFR in complex with L-aspartate | | Descriptor: | ACETATE ION, ASPARTIC ACID, MAGNESIUM ION, ... | | Authors: | Tarnawski, M, Hellweg, L, Bergner, A, Hiblot, J, Leippe, P, Johnsson, K. | | Deposit date: | 2023-04-26 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray structure of the SF-iAspSnFR in complex with L-aspartate
To Be Published
|
|
8PBB
 
 | | CHAPSO treated partial catalytic component (comprising only AnfD & AnfK, lacking AnfG and FeFeco) of iron nitrogenase from Rhodobacter capsulatus | | Descriptor: | FE(8)-S(7) CLUSTER, Nitrogenase iron-iron protein, beta subunit, ... | | Authors: | Schmidt, F.V, Schulz, L, Zarzycki, J, Prinz, S, Erb, T.J, Rebelein, J.G. | | Deposit date: | 2023-06-09 | | Release date: | 2023-10-04 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Structural insights into the iron nitrogenase complex.
Nat.Struct.Mol.Biol., 31, 2024
|
|
