6DCO
 
 | | Bcl-xL complex with Beclin 1 BH3 domain T108D | | Descriptor: | Bcl-2-like protein 1,Bcl-2-like protein 1, Beclin-1, CHLORIDE ION | | Authors: | Lee, E.F, Smith, B.J, Smith, N.A, Yao, S, Fairlie, W.D. | | Deposit date: | 2018-05-07 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Structural insights into BCL2 pro-survival protein interactions with the key autophagy regulator BECN1 following phosphorylation by STK4/MST1.
Autophagy, 15, 2019
|
|
5BY6
 
 | | Crystal structure of Trichinella spiralis thymidylate synthase complexed with dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, GLYCEROL, ... | | Authors: | Dowiercial, A, Jarmula, A, Rypniewski, W, Fraczyk, T, Wilk, P, Rode, W. | | Deposit date: | 2015-06-10 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of nematode (parasitic T. spiralis and free living C. elegans), compared to mammalian, thymidylate synthases (TS). Molecular docking and molecular dynamics simulations in search for nematode-specific inhibitors of TS.
J. Mol. Graph. Model., 77, 2017
|
|
6K4L
 
 | |
5XFV
 
 | | Crystal structures of FMN-bound form of dihydroorotate dehydrogenase from Trypanosoma brucei | | Descriptor: | Dihydroorotate dehydrogenase (fumarate), FLAVIN MONONUCLEOTIDE, MALONATE ION | | Authors: | Kubota, T, Tani, O, Yamaguchi, T, Namatame, I, Sakashita, H, Furukawa, K, Yamasaki, K. | | Deposit date: | 2017-04-11 | | Release date: | 2018-04-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structures of FMN-bound and FMN-free forms of dihydroorotate dehydrogenase fromTrypanosoma brucei.
FEBS Open Bio, 8, 2018
|
|
9GAX
 
 | | TEAD1 in complex with a reversible inhibitor N-[(1S)-2-hydroxy-1-(1-methyl-1H-pyrazol-3-yl)ethyl]-2-methyl-8-[4-(trifluoromethyl)phenyl]-2H,8H-pyrazolo[3,4-b]indole-5-carboxamide | | Descriptor: | 2-methyl-~{N}-[(1~{S})-1-(1-methylpyrazol-3-yl)-2-oxidanyl-ethyl]-4-[4-(trifluoromethyl)phenyl]pyrazolo[3,4-b]indole-7-carboxamide, GLYCEROL, SULFATE ION, ... | | Authors: | Musil, D, Freire, F. | | Deposit date: | 2024-07-29 | | Release date: | 2025-01-22 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | MoA Studies of the TEAD P-Site Binding Ligand MSC-4106 and Its Optimization to TEAD1-Selective Amide M3686.
J.Med.Chem., 68, 2025
|
|
7UG7
 
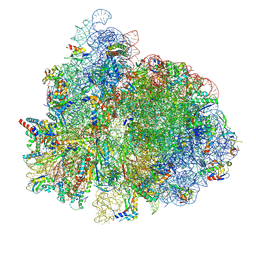 | | 70S ribosome complex in an intermediate state of translocation bound to EF-G(GDP) stalled by Argyrin B | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Rundlet, E.J, Wieland, M, Holm, M, Koller, T.O, Blanchard, S.C, Wilson, D.N. | | Deposit date: | 2022-03-24 | | Release date: | 2022-05-18 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | The cyclic octapeptide antibiotic argyrin B inhibits translation by trapping EF-G on the ribosome during translocation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5K6V
 
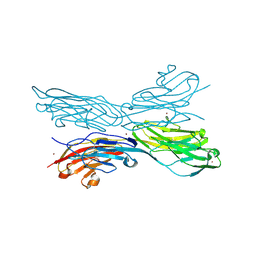 | | Sidekick-1 immunoglobulin domains 1-4, crystal form 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Protein sidekick-1, ... | | Authors: | Jin, X, Goodman, K.M, Mannepalli, S, Honig, B, Shapiro, L. | | Deposit date: | 2016-05-25 | | Release date: | 2016-09-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.208 Å) | | Cite: | Molecular basis of sidekick-mediated cell-cell adhesion and specificity.
Elife, 5, 2016
|
|
5F5L
 
 | |
6DXS
 
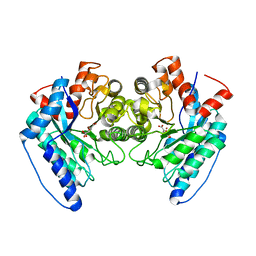 | | Crystal structure of the LigJ hydratase E284Q mutant substrate complex with (3Z)-2-keto-4-carboxy-3-hexenedioate | | Descriptor: | (2Z)-4-oxobut-2-ene-1,2,4-tricarboxylic acid, 4-oxalomesaconate hydratase, ZINC ION | | Authors: | Mabanglo, M.F, Raushel, F.M, Hogancamp, T.N. | | Deposit date: | 2018-06-29 | | Release date: | 2018-09-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and Reaction Mechanism of the LigJ Hydratase: An Enzyme Critical for the Bacterial Degradation of Lignin in the Protocatechuate 4,5-Cleavage Pathway.
Biochemistry, 57, 2018
|
|
7EM6
 
 | | Crystal structure of the PI5P4Kbeta N203D-ITP complex | | Descriptor: | INOSINE-5'-DIPHOSPHATE, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta, [[(2~{R},3~{S},4~{R},5~{R})-3,4-bis(oxidanyl)-5-(6-oxidanylidene-1~{H}-purin-9-yl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Senda, M, Senda, T. | | Deposit date: | 2021-04-13 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The GTP responsiveness of PI5P4K beta evolved from a compromised trade-off between activity and specificity.
Structure, 30, 2022
|
|
6DY1
 
 | | Rabbit N-acylethanolamine-hydrolyzing acid amidase (NAAA) with fatty acid (myristate), in presence of Triton X-100 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{2-[4-(1,1,3,3-TETRAMETHYLBUTYL)PHENOXY]ETHOXY}ETHANOL, CHLORIDE ION, ... | | Authors: | Gorelik, A, Gebai, A, Illes, K, Piomelli, D, Nagar, B. | | Deposit date: | 2018-07-01 | | Release date: | 2018-09-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.998 Å) | | Cite: | Molecular mechanism of activation of the immunoregulatory amidase NAAA.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6AA3
 
 | | Crystal structure of MTH1 in apo form (cocktail No. 1) | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, SULFATE ION, ZINC ION | | Authors: | Yokoyama, T, Kitakami, R, Mizuguchi, M. | | Deposit date: | 2018-07-17 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Discovery of a new class of MTH1 inhibitor by X-ray crystallographic screening.
Eur J Med Chem, 167, 2019
|
|
6TGA
 
 | | Cryo-EM Structure of as isolated form of NAD+-dependent Formate Dehydrogenase from Rhodobacter capsulatus | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Wendler, P, Radon, C, Mittelstaedt, G. | | Deposit date: | 2019-11-15 | | Release date: | 2020-04-22 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Cryo-EM structures reveal intricate Fe-S cluster arrangement and charging in Rhodobacter capsulatus formate dehydrogenase.
Nat Commun, 11, 2020
|
|
9MQV
 
 | |
9MQW
 
 | |
5GKW
 
 | | crystal structure of SZ529 complex with (R,R)-cyclopentanediol | | Descriptor: | (1~{R},2~{R})-cyclopentane-1,2-diol, Limonene-1,2-epoxide hydrolase | | Authors: | Wu, L, Sun, Z.T, Reetz, M.T, Zhou, J.H. | | Deposit date: | 2016-07-07 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | crystal structure of SZ529 complex with (R,R)-cyclopentanediol
To Be Published
|
|
8WTI
 
 | | Crystal structure of the SARS-CoV-2 main protease in complex with 20j | | Descriptor: | 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER, SULFATE ION, ... | | Authors: | Zeng, R, Zhao, X, Yang, S.Y, Lei, J. | | Deposit date: | 2023-10-18 | | Release date: | 2024-08-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of alpha-Ketoamide inhibitors of SARS-CoV-2 main protease derived from quaternized P1 groups.
Bioorg.Chem., 143, 2024
|
|
4QZ3
 
 | | yCP beta5-A49V mutant in complex with the epoxyketone inhibitor ONX 0914 | | Descriptor: | 1,2,4-trideoxy-4-methyl-2-{[N-(morpholin-4-ylacetyl)-L-alanyl-O-methyl-L-tyrosyl]amino}-1-phenyl-D-xylitol, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Huber, E.M, Heinemeyer, W, Groll, M. | | Deposit date: | 2014-07-27 | | Release date: | 2015-02-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Bortezomib-Resistant Mutant Proteasomes: Structural and Biochemical Evaluation with Carfilzomib and ONX 0914.
Structure, 23, 2015
|
|
5XT4
 
 | | Crystal Structure of Transketolase in complex with TPP intermediate VIII' from Pichia Stipitis | | Descriptor: | 2-C-{3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-5-(2-{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-4-methyl-1,3-thiazol-3-ium-2-yl}-6-O-phosphono-D-glucitol, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Li, T.L, Hsu, N.S, Wang, Y.L. | | Deposit date: | 2017-06-16 | | Release date: | 2018-06-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Evidence of Diradicals Involved in the Yeast Transketolase Catalyzed Keto-Transferring Reactions.
Chembiochem, 19, 2018
|
|
5XSW
 
 | |
4Z0P
 
 | | Crystal structure of NADPH-dependent glyoxylate/hydroxypyruvate reductase SMc02828 (SmGhrA) from Sinorhizobium meliloti in complex with NADPH and oxalate | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | Sroka, P, Gasiorowska, O.A, Handing, K.B, Shabalin, I.G, Porebski, P.J, Hillerich, B.S, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-03-26 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural, Biochemical, and Evolutionary Characterizations of Glyoxylate/Hydroxypyruvate Reductases Show Their Division into Two Distinct Subfamilies.
Biochemistry, 57, 2018
|
|
7BW2
 
 | | Crystal Structure of Cyanobacterial PSI Monomer from T.elongatus at 6.5 A Resolution | | Descriptor: | Photosystem I 4.8K protein, Photosystem I P700 chlorophyll a apoprotein A1, Photosystem I P700 chlorophyll a apoprotein A2, ... | | Authors: | Kurisu, G, Coruh, O, Tanaka, H, Eithar, E.M, Mian, Y. | | Deposit date: | 2020-04-13 | | Release date: | 2021-03-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (6.5 Å) | | Cite: | Cryo-EM structure of a functional monomeric Photosystem I from Thermosynechococcus elongatus reveals red chlorophyll cluster.
Commun Biol, 4, 2021
|
|
9RTI
 
 | |
4QZ4
 
 | | yCP beta5-A49S mutant in complex with the epoxyketone inhibitor ONX 0914 | | Descriptor: | 1,2,4-trideoxy-4-methyl-2-{[N-(morpholin-4-ylacetyl)-L-alanyl-O-methyl-L-tyrosyl]amino}-1-phenyl-D-xylitol, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Huber, E.M, Heinemeyer, W, Groll, M. | | Deposit date: | 2014-07-27 | | Release date: | 2015-02-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Bortezomib-Resistant Mutant Proteasomes: Structural and Biochemical Evaluation with Carfilzomib and ONX 0914.
Structure, 23, 2015
|
|
5CI0
 
 | | Ribonucleotide reductase Y122 3,5-F2Y variant | | Descriptor: | MU-OXO-DIIRON, Ribonucleoside-diphosphate reductase 1, beta subunit, ... | | Authors: | Funk, M.A, Drennan, C.L. | | Deposit date: | 2015-07-10 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Biophysical Characterization of Fluorotyrosine Probes Site-Specifically Incorporated into Enzymes: E. coli Ribonucleotide Reductase As an Example.
J.Am.Chem.Soc., 138, 2016
|
|
