6O4Z
 
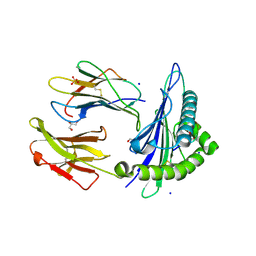 | | Structure of HLA-A2:01 with peptide MM92 | | Descriptor: | Beta-2-microglobulin, GLYCEROL, MHC class I antigen, ... | | Authors: | Ying, G, Bitra, A, Zajonc, D.M. | | Deposit date: | 2019-03-01 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Anin silico-in vitroPipeline Identifying an HLA-A*02:01+KRAS G12V+Spliced Epitope Candidate for a Broad Tumor-Immune Response in Cancer Patients.
Front Immunol, 10, 2019
|
|
6O51
 
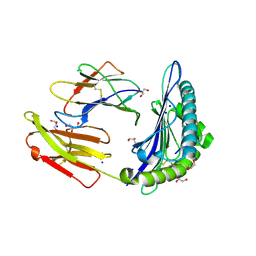 | | Structure of HLA-A2:01 with peptide MM90 | | Descriptor: | Beta-2-microglobulin, GLYCEROL, MHC class I antigen, ... | | Authors: | Ying, G, Bitra, A, Zajonc, D.M. | | Deposit date: | 2019-03-01 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Anin silico-in vitroPipeline Identifying an HLA-A*02:01+KRAS G12V+Spliced Epitope Candidate for a Broad Tumor-Immune Response in Cancer Patients.
Front Immunol, 10, 2019
|
|
8UVR
 
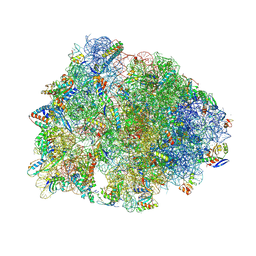 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with spectinomycin, mRNA, deacylated A- and E-site tRNAphe, and deacylated P-site tRNAmet at 2.60A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Killam, B.Y, Phelps, G.A, Lee, R.E, Polikanov, Y.S. | | Deposit date: | 2023-11-03 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Development of 2nd generation aminomethyl spectinomycins that overcome native efflux in Mycobacterium abscessus.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8J30
 
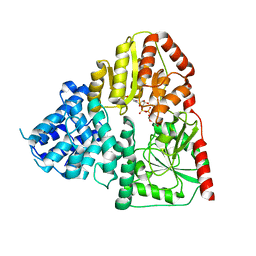 | | Crystal structure of ApNGT with Q469A and M218A mutations in complex with UDP-GLC | | Descriptor: | UDP-glucose:protein N-beta-glucosyltransferase, URIDINE-5'-DIPHOSPHATE, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Feng, Y, Hao, Z, Guo, Q, Zheng, J, Da, L, Peng, W. | | Deposit date: | 2023-04-15 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Investigation of the Catalytic Mechanism of a Soluble N-glycosyltransferase Allows Synthesis of N-glycans at Noncanonical Sequons.
Jacs Au, 3, 2023
|
|
7UG7
 
 | | 70S ribosome complex in an intermediate state of translocation bound to EF-G(GDP) stalled by Argyrin B | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Rundlet, E.J, Wieland, M, Holm, M, Koller, T.O, Blanchard, S.C, Wilson, D.N. | | Deposit date: | 2022-03-24 | | Release date: | 2022-05-18 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | The cyclic octapeptide antibiotic argyrin B inhibits translation by trapping EF-G on the ribosome during translocation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6OBV
 
 | | Structural insights into dehydratase substrate selection for the borrelidin and fluvirucin polyketide synthases | | Descriptor: | fluvirucin B1 DH domain from module 1 | | Authors: | McAndrew, R.P, Barajas, J.F, Pereira, J.H, Keasling, J.D, Adams, P.D. | | Deposit date: | 2019-03-21 | | Release date: | 2019-06-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural insights into dehydratase substrate selection for the borrelidin and fluvirucin polyketide synthases.
J Ind Microbiol Biotechnol., 46, 2019
|
|
8K5S
 
 | |
8K5T
 
 | |
3JC2
 
 | |
2A02
 
 | | Solution NMR Structure of the Periplasmic Signaling Domain of the Outer Membrane Iron Transporter PupA from Pseudomonas putida. | | Descriptor: | Ferric-pseudobactin 358 receptor | | Authors: | Ferguson, A.D, Amezcua, C.A, Chelliah, Y, Rosen, M.K, Deisenhofer, J. | | Deposit date: | 2005-06-15 | | Release date: | 2006-09-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Signal transduction pathway of TonB-dependent transporters.
Proc.Natl.Acad.Sci.Usa, 2, 2006
|
|
3HLF
 
 | | Simvastatin Synthase (LovD), from Aspergillus terreus, S5 mutant, S76A mutant, complex with simvastatin | | Descriptor: | Simvastatin acid, Transesterase | | Authors: | Sawaya, M.R, Yeates, T.O, Pashkov, I, Gao, X, Tang, Y. | | Deposit date: | 2009-05-27 | | Release date: | 2009-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Directed evolution and structural characterization of a simvastatin synthase
Chem.Biol., 16, 2009
|
|
4NXQ
 
 | | Crystal Structure of T-cell Lymphoma Invasion and Metastasis-1 PDZ Domain Quadruple Mutant (QM) in Complex With Caspr4 Peptide | | Descriptor: | Contactin-associated protein-like 4 peptide, T-lymphoma invasion and metastasis-inducing protein 1 | | Authors: | Liu, X, Speckhard, D.C, Shepherd, T.R, Hengel, S.R, Fuentes, E.J. | | Deposit date: | 2013-12-09 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Distinct Roles for Conformational Dynamics in Protein-Ligand Interactions.
Structure, 24, 2016
|
|
6UH3
 
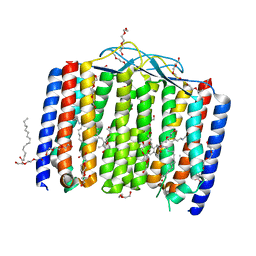 | | Crystal structure of bacterial heliorhodopsin 48C12 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Heliorhodopsin, PALMITIC ACID, ... | | Authors: | Lu, Y, Zhou, X.E, Gao, X, Xia, R, Xu, Z, Wang, N, Leng, Y, Melcher, K, Xu, H.E, He, Y. | | Deposit date: | 2019-09-26 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of heliorhodopsin 48C12.
Cell Res., 30, 2020
|
|
6UVQ
 
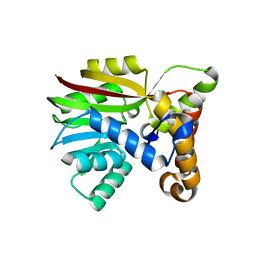 | | Crystal structure of Apo AtmM | | Descriptor: | ACETATE ION, D-glucose O-methyltransferase, MAGNESIUM ION | | Authors: | Alvarado, S.K, Wang, Z, Miller, M.D, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2019-11-04 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structure of Apo AtmM
To Be Published
|
|
6UWD
 
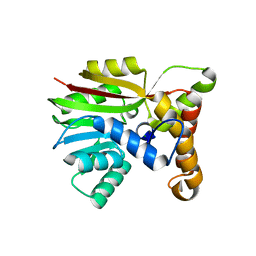 | | Crystal structure of Apo AtmM | | Descriptor: | ACETATE ION, D-glucose O-methyltransferase, MAGNESIUM ION | | Authors: | Alvarado, S.K, Wang, Z, Miller, M.D, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2019-11-05 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure of Apo AtmM
To Be Published
|
|
4XWF
 
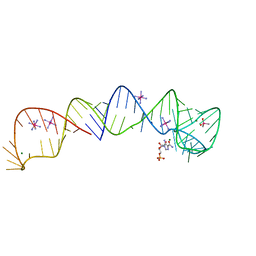 | |
4XW7
 
 | |
8FHJ
 
 | | Crystal structure of a FAD monooxygenease from Methylocystis sp. Strain SB2 | | Descriptor: | BROMIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Stewart, A.M, Sawaya, M.R, Stewart, C.E. | | Deposit date: | 2022-12-14 | | Release date: | 2023-05-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal structure of MbnF: an NADPH-dependent flavin monooxygenase from Methylocystis strain SB2.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
6UV6
 
 | | AtmM with bound rebeccamycin analogue | | Descriptor: | 12-beta-D-glucopyranosyl-12,13-dihydro-5H-indolo[2,3-a]pyrrolo[3,4-c]carbazole-5,7(6H)-dione, D-glucose O-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Alvarado, S.K, Wang, Z, Miller, M.D, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2019-11-01 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structure of AtmM Bound with Glycosylated Indolocarbazole
To Be Published
|
|
6WB7
 
 | | Acarbose Kinase AcbK as a Complex with Acarbose and AMP-PNP | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Acarbose 7(IV)-phosphotransferase, MANGANESE (II) ION, ... | | Authors: | Jeffrey, P.D, Balaich, J.N, Estrella, M.A, Donia, M.S. | | Deposit date: | 2020-03-26 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | The human microbiome encodes resistance to the antidiabetic drug acarbose.
Nature, 600, 2021
|
|
7WIV
 
 | | Cryo-EM structure of Mycobacterium tuberculosis irtAB in complex with an AMP-PNP | | Descriptor: | Mycobactin import ATP-binding/permease protein IrtA, Mycobactin import ATP-binding/permease protein IrtB, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Zhang, B, Sun, S, Yang, H, Rao, Z. | | Deposit date: | 2022-01-05 | | Release date: | 2023-01-25 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Cryo-EM structures for the Mycobacterium tuberculosis iron-loaded siderophore transporter IrtAB.
Protein Cell, 14, 2023
|
|
7WIU
 
 | | Cryo-EM structure of Mycobacterium tuberculosis irtAB in inward-facing state | | Descriptor: | Mycobactin import ATP-binding/permease protein IrtA, Mycobactin import ATP-binding/permease protein IrtB | | Authors: | Zhang, B, Sun, S, Yang, H, Rao, Z. | | Deposit date: | 2022-01-05 | | Release date: | 2023-01-25 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Cryo-EM structures for the Mycobacterium tuberculosis iron-loaded siderophore transporter IrtAB.
Protein Cell, 14, 2023
|
|
8GJP
 
 | |
8GJ4
 
 | |
8GKM
 
 | |
