5C5S
 
 | |
8JM9
 
 | |
8JMI
 
 | |
8GQ9
 
 | | Crystal structure of lasso peptide epimerase MslH | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Nakashima, Y, Morita, H. | | Deposit date: | 2022-08-29 | | Release date: | 2023-06-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of lasso peptide epimerase MslH reveals metal-dependent acid/base catalytic mechanism.
Nat Commun, 14, 2023
|
|
8GQA
 
 | |
8JMA
 
 | |
8JME
 
 | |
8JMH
 
 | |
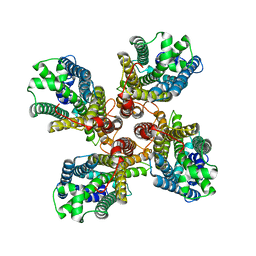
8GQB
 
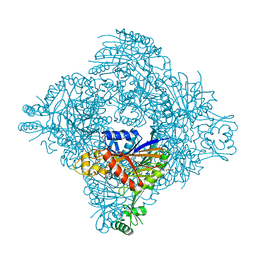 | |
6P12
 
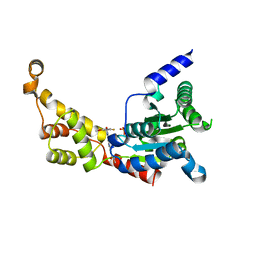 | | Structure of spastin AAA domain (wild-type) in complex with diaminotriazole-based inhibitor | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-{[5-amino-1-(2-fluoro-6-methoxybenzene-1-carbonyl)-1H-1,2,4-triazol-3-yl]amino}-N-methylbenzamide, Drosophila melanogaster Spastin AAA domain, ... | | Authors: | Pisa, R, Cupido, T, Kapoor, T.M. | | Deposit date: | 2019-05-17 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.94131851 Å) | | Cite: | Analyzing Resistance to Design Selective Chemical Inhibitors for AAA Proteins.
Cell Chem Biol, 26, 2019
|
|
5MWE
 
 | | Complex between the Leucine Zipper (LZ, residues 490-567) and Centrosomin-motif 2 (CM2) domains of Drosophila melanogaster Centrosomin (Cnn) | | Descriptor: | 1,2-ETHANEDIOL, 3,3',3''-phosphanetriyltripropanoic acid, Centrosomin, ... | | Authors: | Feng, Z, Johnson, S, Raff, J.W, Lea, S.M. | | Deposit date: | 2017-01-18 | | Release date: | 2017-06-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural Basis for Mitotic Centrosome Assembly in Flies.
Cell, 169, 2017
|
|
4Y28
 
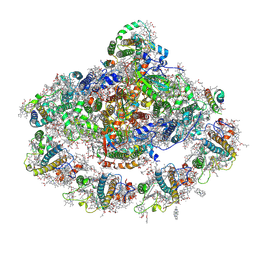 | | The structure of plant photosystem I super-complex at 2.8 angstrom resolution. | | Descriptor: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Mazor, Y, Brovikov, A, Nelson, N. | | Deposit date: | 2015-02-09 | | Release date: | 2015-08-19 | | Last modified: | 2019-04-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structure of plant photosystem I super-complex at 2.8 angstrom resolution.
Elife, 4, 2015
|
|
5MW9
 
 | | Complex between the Leucine Zipper (LZ) and Centrosomin-motif 2 (CM2) domains of Drosophila melanogaster Centrosomin (Cnn) - L535E mutant form | | Descriptor: | Centrosomin, ZINC ION | | Authors: | Feng, Z, Johnson, S, Raff, J.W, Lea, S.M. | | Deposit date: | 2017-01-18 | | Release date: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Mitotic Centrosome Assembly in Flies.
Cell, 169, 2017
|
|
5MVW
 
 | | Complex between the Leucine Zipper (LZ) and Centrosomin-motif 2 (CM2) domains of Drosophila melanogaster Centrosomin (Cnn) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Centrosomin, ... | | Authors: | Feng, Z, Johnson, S, Raff, J.W, Lea, S.M. | | Deposit date: | 2017-01-17 | | Release date: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural Basis for Mitotic Centrosome Assembly in Flies.
Cell, 169, 2017
|
|
5MW0
 
 | | Complex between the Leucine Zipper (LZ) and Centrosomin-motif 2 (CM2) domains of Drosophila melanogaster Centrosomin (Cnn) - L535E mutant form | | Descriptor: | Centrosomin, ZINC ION | | Authors: | Feng, Z, Johnson, S, Raff, J.W, Lea, S.M. | | Deposit date: | 2017-01-17 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Mitotic Centrosome Assembly in Flies.
Cell, 169, 2017
|
|
7BGI
 
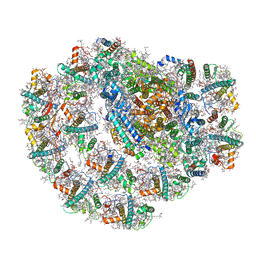 | | Photosystem I of a temperature sensitive mutant Chlamydomonas reinhardtii | | Descriptor: | (1~{S})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(4~{S})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15,17-nonaenyl]cyclohex-3-en-1-ol, (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate, (2S)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-hydroxypropyl hexadecanoate, ... | | Authors: | Caspy, I, Nelson, N. | | Deposit date: | 2021-01-07 | | Release date: | 2021-12-08 | | Last modified: | 2021-12-15 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Dimeric and high-resolution structures of Chlamydomonas Photosystem I from a temperature-sensitive Photosystem II mutant
Commun Biol, 4, 2021
|
|
5JVS
 
 | |
7BLX
 
 | | Photosystem I of a temperature sensitive mutant Chlamydomonas reinhardtii | | Descriptor: | (1~{S})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(4~{S})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15,17-nonaenyl]cyclohex-3-en-1-ol, (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate, (2S)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-hydroxypropyl hexadecanoate, ... | | Authors: | Caspy, I, Nelson, N. | | Deposit date: | 2021-01-19 | | Release date: | 2021-12-15 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Dimeric and high-resolution structures of Chlamydomonas Photosystem I from a temperature-sensitive Photosystem II mutant
Commun Biol, 4, 2021
|
|
5JVU
 
 | |
6VO1
 
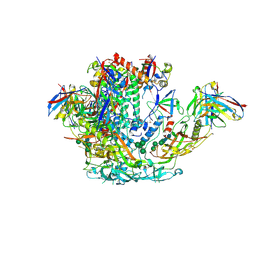 | | BG505 SOSIP.v5.2 in complex with rhesus macaque Fab RM20J | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Cottrell, C.A, Ward, A.B. | | Deposit date: | 2020-01-29 | | Release date: | 2020-07-01 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Mapping the immunogenic landscape of near-native HIV-1 envelope trimers in non-human primates.
Plos Pathog., 16, 2020
|
|
5JVR
 
 | |
5GTI
 
 | | Native XFEL structure of photosystem II (two flash dataset) | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Shen, J.R. | | Deposit date: | 2016-08-20 | | Release date: | 2017-03-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Light-induced structural changes and the site of O=O bond formation in PSII caught by XFEL.
Nature, 543, 2017
|
|
5H2F
 
 | | Crystal structure of the PsbM-deletion mutant of photosystem II | | Descriptor: | (3R)-beta,beta-caroten-3-ol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Uto, S, Kawakami, K, Umena, Y, Iwai, M, Ikeuchi, M, Shen, J.R, Kamiya, N. | | Deposit date: | 2016-10-15 | | Release date: | 2017-03-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mutual relationships between structural and functional changes in a PsbM-deletion mutant of photosystem II.
Faraday Discuss., 198, 2017
|
|
5GTH
 
 | | Native XFEL structure of photosystem II (dark dataset) | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Shen, J.R. | | Deposit date: | 2016-08-20 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Light-induced structural changes and the site of O=O bond formation in PSII caught by XFEL.
Nature, 543, 2017
|
|
8H3W
 
 | |
