9CSS
 
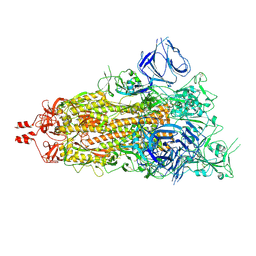 | | Cryo-EM structure of SARS-CoV-2 spike protein Ecto-domain with internal tag, 1UP RBD conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Singh, S, Hasan, S.S. | | Deposit date: | 2024-07-24 | | Release date: | 2025-03-05 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Production and cryo-electron microscopy structure of an internally tagged SARS-CoV-2 spike ecto-domain construct.
J Struct Biol X, 11, 2025
|
|
9DO1
 
 | | SARS-CoV-2 papain-like protease (PLpro) with inhibitor Jun13307 | | Descriptor: | 2-methyl-N-{(1R)-1-[(2M)-2-(1-methyl-1H-pyrazol-4-yl)quinolin-4-yl]ethyl}-5-{[(2R)-1-methylpyrrolidin-2-yl]methoxy}benzamide, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ansari, A, Jadhav, P, Arnold, E, Ruiz, F.X, Wang, J. | | Deposit date: | 2024-09-18 | | Release date: | 2025-01-22 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design of quinoline SARS-CoV-2 papain-like protease inhibitors as oral antiviral drug candidates.
Nat Commun, 16, 2025
|
|
9DO3
 
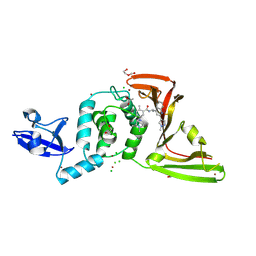 | | SARS-CoV-2 papain-like protease (PLpro) with inhibitor Jun13317 | | Descriptor: | 1,2-ETHANEDIOL, 5-[(3S)-3,4-dimethylpiperazin-1-yl]-2-methyl-N-{(1R)-1-[(2M)-2-(1-methyl-1H-pyrazol-4-yl)quinolin-4-yl]ethyl}benzamide, ACETATE ION, ... | | Authors: | Ansari, A, Jadhav, P, Arnold, E, Ruiz, F.X, Wang, J. | | Deposit date: | 2024-09-18 | | Release date: | 2025-01-22 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design of quinoline SARS-CoV-2 papain-like protease inhibitors as oral antiviral drug candidates.
Nat Commun, 16, 2025
|
|
9BD9
 
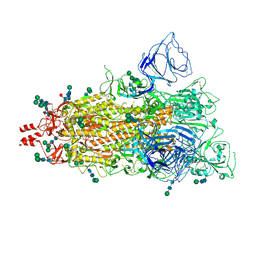 | | SARS CoV-2 full-length WT spike protein, 1RBD-up conformation (SPIKE-WT) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Singh, S, Hasan, S.S. | | Deposit date: | 2024-04-11 | | Release date: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structure of SARS CoV-2 full-length spike protein, 1RBD-up conformation
To Be Published
|
|
9B8F
 
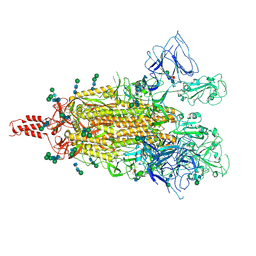 | |
9BBK
 
 | |
9DNU
 
 | | SARS-CoV-2 papain-like protease (PLpro) with inhibitor Jun13296 | | Descriptor: | 1,2-ETHANEDIOL, 2-methyl-5-[(1R,5S)-8-methyl-3,8-diazabicyclo[3.2.1]octan-3-yl]-N-{(1R)-1-[(2P)-2-(1-methyl-1H-pyrazol-4-yl)quinolin-4-yl]ethyl}benzamide, ACETATE ION, ... | | Authors: | Ansari, A, Jadhav, P, Arnold, E, Ruiz, F.X, Wang, J. | | Deposit date: | 2024-09-18 | | Release date: | 2025-01-22 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design of quinoline SARS-CoV-2 papain-like protease inhibitors as oral antiviral drug candidates.
Nat Commun, 16, 2025
|
|
9DOI
 
 | | SARS-CoV-2 papain-like protease (PLpro) with inhibitor Jun13306 | | Descriptor: | 2-methyl-N-{(1S)-1-[(2P)-2-(1-methyl-1H-pyrazol-4-yl)quinolin-4-yl]ethyl}-5-{[(2S)-1-methylpyrrolidin-2-yl]methoxy}benzamide, ACETATE ION, CHLORIDE ION, ... | | Authors: | Ansari, A, Jadhav, P, Arnold, E, Ruiz, F.X, Wang, J. | | Deposit date: | 2024-09-19 | | Release date: | 2025-01-22 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design of quinoline SARS-CoV-2 papain-like protease inhibitors as oral antiviral drug candidates.
Nat Commun, 16, 2025
|
|
9DO5
 
 | | SARS-CoV-2 papain-like protease (PLpro) with inhibitor Jun12665 | | Descriptor: | 5-[2-(dimethylamino)ethoxy]-N-{(1R)-1-[(2P)-2-{1-[2-(dimethylamino)-2-oxoethyl]-1H-pyrazol-4-yl}quinolin-4-yl]ethyl}-2-methylbenzamide, ACETATE ION, CHLORIDE ION, ... | | Authors: | Ansari, A, Jadhav, P, Arnold, E, Ruiz, F.X, Wang, J. | | Deposit date: | 2024-09-18 | | Release date: | 2025-01-22 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Design of quinoline SARS-CoV-2 papain-like protease inhibitors as oral antiviral drug candidates.
Nat Commun, 16, 2025
|
|
6XF5
 
 | | Cryo-EM structure of a biotinylated SARS-CoV-2 spike probe in the prefusion state (RBDs down) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Cerutti, G, Gorman, J, Kwong, P.D, Shapiro, L. | | Deposit date: | 2020-06-15 | | Release date: | 2020-09-02 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structure-Based Design with Tag-Based Purification and In-Process Biotinylation Enable Streamlined Development of SARS-CoV-2 Spike Molecular Probes.
SSRN, 2020
|
|
6XCN
 
 | |
6XF6
 
 | | Cryo-EM structure of a biotinylated SARS-CoV-2 spike probe in the prefusion state (1 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Cerutti, G, Gorman, J, Kwong, P.D, Shapiro, L. | | Deposit date: | 2020-06-15 | | Release date: | 2020-09-02 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure-Based Design with Tag-Based Purification and In-Process Biotinylation Enable Streamlined Development of SARS-CoV-2 Spike Molecular Probes.
SSRN, 2020
|
|
6WOJ
 
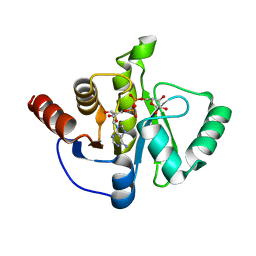 | | Structure of the SARS-CoV-2 macrodomain (NSP3) in complex with ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Non-structural protein 3 | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Gao, F.P, Fehr, A.R. | | Deposit date: | 2020-04-24 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The SARS-CoV-2 Conserved Macrodomain Is a Mono-ADP-Ribosylhydrolase.
J.Virol., 95, 2021
|
|
6XMK
 
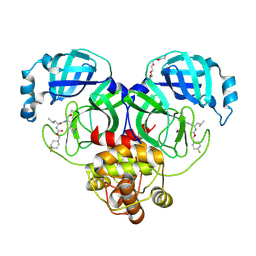 | | 1.70 A resolution structure of SARS-CoV-2 3CL protease in complex with inhibitor 7j | | Descriptor: | (1S,2S)-2-[(N-{[(4,4-difluorocyclohexyl)methoxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, TETRAETHYLENE GLYCOL | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Rathnayake, A.D, Zheng, J, Kim, Y, Nguyen, H.N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2020-06-30 | | Release date: | 2020-07-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 3C-like protease inhibitors block coronavirus replication in vitro and improve survival in MERS-CoV-infected mice.
Sci Transl Med, 12, 2020
|
|
6XCM
 
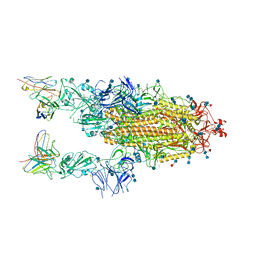 | |
6X79
 
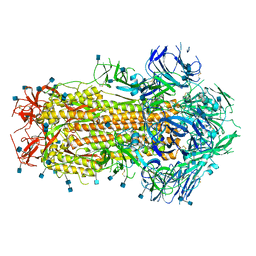 | | Prefusion SARS-CoV-2 S ectodomain trimer covalently stabilized in the closed conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | McCallum, M, Walls, A.C, Corti, D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2020-05-29 | | Release date: | 2020-08-19 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure-guided covalent stabilization of coronavirus spike glycoprotein trimers in the closed conformation.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7JMW
 
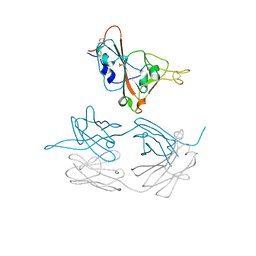 | | Crystal structure of SARS-CoV-2 spike protein receptor-binding domain in complex with cross-neutralizing antibody COVA1-16 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COVA1-16 heavy chain, COVA1-16 light chain, ... | | Authors: | Liu, H, Yuan, M, Zhu, X, Wu, N.C, Wilson, I.A. | | Deposit date: | 2020-08-03 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Cross-Neutralization of a SARS-CoV-2 Antibody to a Functionally Conserved Site Is Mediated by Avidity.
Immunity, 53, 2020
|
|
7JV6
 
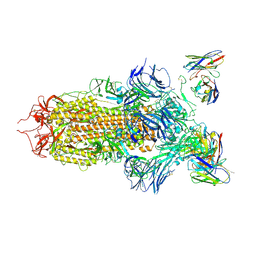 | | SARS-CoV-2 spike in complex with the S2H13 neutralizing antibody (closed conformation) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S2H13 Fab heavy chain, ... | | Authors: | Park, Y.J, Tortorici, M.A, Walls, A.C, Czudnochowski, N, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Snell, G, Veesler, D. | | Deposit date: | 2020-08-20 | | Release date: | 2020-10-14 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.
Cell, 183, 2020
|
|
7UHC
 
 | |
7JTL
 
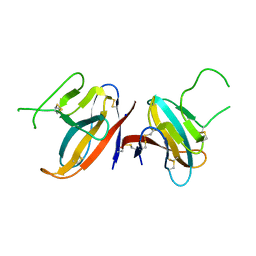 | | Structure of SARS-CoV-2 ORF8 accessory protein | | Descriptor: | ORF8 protein, SODIUM ION | | Authors: | Flower, T.G, Buffalo, C.Z, Hooy, R.M, Allaire, M, Ren, X, Hurley, J.H. | | Deposit date: | 2020-08-18 | | Release date: | 2020-08-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure of SARS-CoV-2 ORF8, a rapidly evolving immune evasion protein.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7JVA
 
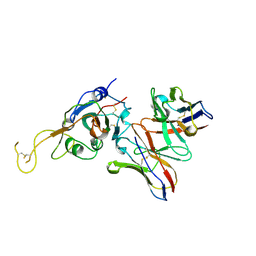 | | SARS-CoV-2 spike in complex with the S2A4 neutralizing antibody Fab fragment (local refinement of the receptor-binding domain and Fab variable domains) | | Descriptor: | S2A4 Fab heavy chain, S2A4 Fab light chain, Spike glycoprotein, ... | | Authors: | Park, Y.J, Tortorici, M.A, Walls, A.C, Czudnochowski, N, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Snell, G, Veesler, D. | | Deposit date: | 2020-08-20 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.
Cell, 183, 2020
|
|
7JMX
 
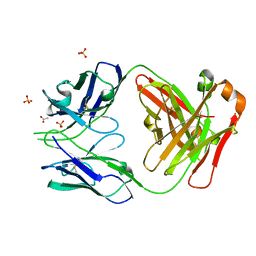 | | Crystal structure of a SARS-CoV-2 cross-neutralizing antibody COVA1-16 Fab | | Descriptor: | ACETATE ION, COVA1-16 heavy chain, COVA1-16 light chain, ... | | Authors: | Liu, H, Yuan, M, Zhu, X, Wu, N.C, Wilson, I.A. | | Deposit date: | 2020-08-03 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Cross-Neutralization of a SARS-CoV-2 Antibody to a Functionally Conserved Site Is Mediated by Avidity.
Immunity, 53, 2020
|
|
7JV2
 
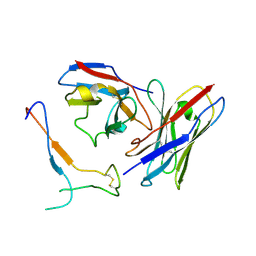 | | SARS-CoV-2 spike in complex with the S2H13 neutralizing antibody Fab fragment (local refinement of the receptor-binding motif and Fab variable domains) | | Descriptor: | S2H13 Fab heavy chain, S2H13 Fab light chain, Spike glycoprotein | | Authors: | Park, Y.J, Tortorici, M.A, Walls, A.C, Czudnochowski, N, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Snell, G, Veesler, D. | | Deposit date: | 2020-08-20 | | Release date: | 2020-10-14 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.
Cell, 183, 2020
|
|
7JW0
 
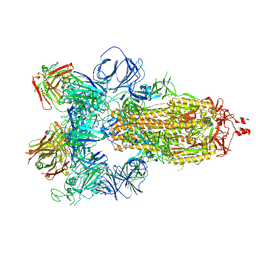 | | SARS-CoV-2 spike in complex with the S304 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S304 Fab heavy chain, ... | | Authors: | Walls, A.C, Park, Y.J, Tortorici, M.A, Czudnochowski, N, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Snell, G, Veesler, D. | | Deposit date: | 2020-08-24 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.
Cell, 183, 2020
|
|
7JVZ
 
 | |
