2GLS
 
 | |
1ALC
 
 | |
2CRO
 
 | |
1GOX
 
 | |
1HNE
 
 | | Structure of human neutrophil elastase in complex with a peptide chloromethyl ketone inhibitor at 1.84-angstroms resolution | | Descriptor: | HUMAN LEUCOCYTE ELASTASE, METHOXYSUCCINYL-ALA-ALA-PRO-ALA CHLOROMETHYL KETONE INHIBITOR | | Authors: | Navia, M.A, Mckeever, B.M, Springer, J.P, Lin, T.-Y, Williams, H.R, Fluder, E.M, Dorn, C.P, Hoogsteen, K. | | Deposit date: | 1989-04-10 | | Release date: | 1989-10-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structure of human neutrophil elastase in complex with a peptide chloromethyl ketone inhibitor at 1.84-A resolution.
Proc.Natl.Acad.Sci.USA, 86, 1989
|
|
2TS1
 
 | |
2UTG
 
 | |
3HVP
 
 | |
2AAT
 
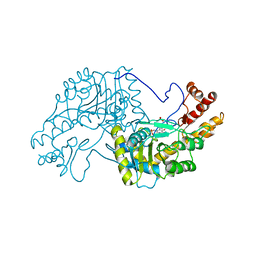 | | 2.8-ANGSTROMS-RESOLUTION CRYSTAL STRUCTURE OF AN ACTIVE-SITE MUTANT OF ASPARTATE AMINOTRANSFERASE FROM ESCHERICHIA COLI | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, ASPARTATE AMINOTRANSFERASE, SULFATE ION | | Authors: | Smith, D, Almo, S.C, Toney, M, Ringe, D. | | Deposit date: | 1989-05-30 | | Release date: | 1989-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | 2.8-A-resolution crystal structure of an active-site mutant of aspartate aminotransferase from Escherichia coli.
Biochemistry, 28, 1989
|
|
2GD1
 
 | |
1UTG
 
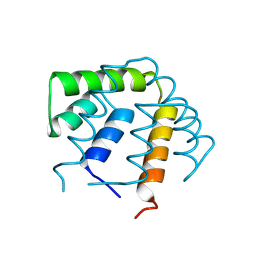 | | REFINEMENT OF THE C2221 CRYSTAL FORM OF OXIDIZED UTEROGLOBIN AT 1.34 ANGSTROMS RESOLUTION | | Descriptor: | UTEROGLOBIN | | Authors: | Morize, I, Surcouf, E, Vaney, M.C, Buehner, M, Mornon, J.P. | | Deposit date: | 1989-04-03 | | Release date: | 1989-10-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Refinement of the C222(1) crystal form of oxidized uteroglobin at 1.34 A resolution.
J.Mol.Biol., 194, 1987
|
|
1R69
 
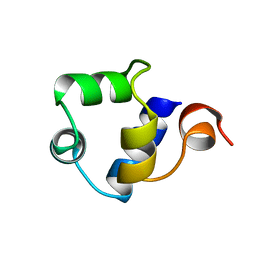 | | STRUCTURE OF THE AMINO-TERMINAL DOMAIN OF PHAGE 434 REPRESSOR AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | REPRESSOR PROTEIN CI | | Authors: | Mondragon, A, Subbiah, S, Alamo, S.C, Drottar, M, Harrison, S.C. | | Deposit date: | 1988-12-08 | | Release date: | 1989-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the amino-terminal domain of phage 434 repressor at 2.0 A resolution.
J.Mol.Biol., 205, 1989
|
|
3RNT
 
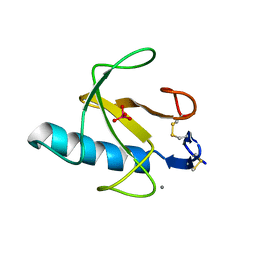 | | CRYSTAL STRUCTURE OF GUANOSINE-FREE RIBONUCLEASE T1, COMPLEXED WITH VANADATE(V), SUGGESTS CONFORMATIONAL CHANGE UPON SUBSTRATE BINDING | | Descriptor: | CALCIUM ION, RIBONUCLEASE T1, VANADATE ION | | Authors: | Kostrewa, D, Choe, H.-W, Heinemann, U, Saenger, W. | | Deposit date: | 1989-05-31 | | Release date: | 1989-10-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of guanosine-free ribonuclease T1, complexed with vanadate (V), suggests conformational change upon substrate binding.
Biochemistry, 28, 1989
|
|
2RSP
 
 | |
2PLV
 
 | |
1I1B
 
 | |
1TLD
 
 | | CRYSTAL STRUCTURE OF BOVINE BETA-TRYPSIN AT 1.5 ANGSTROMS RESOLUTION IN A CRYSTAL FORM WITH LOW MOLECULAR PACKING DENSITY. ACTIVE SITE GEOMETRY, ION PAIRS AND SOLVENT STRUCTURE | | Descriptor: | BETA-TRYPSIN, CALCIUM ION, SULFATE ION | | Authors: | Bartunik, H.D, Summers, L.J, Bartsch, H.H. | | Deposit date: | 1989-07-24 | | Release date: | 1990-01-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of bovine beta-trypsin at 1.5 A resolution in a crystal form with low molecular packing density. Active site geometry, ion pairs and solvent structure.
J.Mol.Biol., 210, 1989
|
|
2RMU
 
 | | THREE-DIMENSIONAL STRUCTURES OF DRUG-RESISTANT MUTANTS OF HUMAN RHINOVIRUS 14 | | Descriptor: | HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP1), HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP2), HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP3), ... | | Authors: | Badger, J, Krishnaswamy, S, Kremer, M.J, Oliveira, M.A, Rossmann, M.G, Heinz, B.A, Rueckert, R.R, Dutko, F.J, Mckinlay, M.A. | | Deposit date: | 1988-10-03 | | Release date: | 1990-01-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Three-dimensional structures of drug-resistant mutants of human rhinovirus 14.
J.Mol.Biol., 207, 1989
|
|
3CA2
 
 | | CRYSTALLOGRAPHIC STUDIES OF INHIBITOR BINDING SITES IN HUMAN CARBONIC ANHYDRASE II. A PENTACOORDINATED BINDING OF THE SCN-ION TO THE ZINC AT HIGH P*H | | Descriptor: | 3-MERCURI-4-AMINOBENZENESULFONAMIDE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Eriksson, A.E, Jones, T.A, Liljas, A. | | Deposit date: | 1989-10-02 | | Release date: | 1990-01-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic studies of inhibitor binding sites in human carbonic anhydrase II: a pentacoordinated binding of the SCN- ion to the zinc at high pH.
Proteins, 4, 1988
|
|
2R04
 
 | | STRUCTURAL ANALYSIS OF ANTIVIRAL AGENTS THAT INTERACT WITH THE CAPSID OF HUMAN RHINOVIRUSES | | Descriptor: | 5-(7-(4-(4,5-DIHYDRO-2-OXAZOLYL)PHENOXY)HEPTYL)-3-METHYL ISOXAZOLE, HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP1), HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP2), ... | | Authors: | Badger, J, Smith, T.J, Rossmann, M.G. | | Deposit date: | 1988-10-03 | | Release date: | 1990-01-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural analysis of antiviral agents that interact with the capsid of human rhinoviruses.
Proteins, 6, 1989
|
|
2R06
 
 | | STRUCTURAL ANALYSIS OF ANTIVIRAL AGENTS THAT INTERACT WITH THE CAPSID OF HUMAN RHINOVIRUSES | | Descriptor: | 5-(5-(4-(4,5-DIHYDRO-2-OXAZOLY)PHENOXY)PENTYL)-3-METHYL ISOXAZOLE, HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP1), HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP2), ... | | Authors: | Badger, J, Smith, T.J, Rossmann, M.G. | | Deposit date: | 1988-10-03 | | Release date: | 1990-01-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural analysis of antiviral agents that interact with the capsid of human rhinoviruses.
Proteins, 6, 1989
|
|
2R07
 
 | | STRUCTURAL ANALYSIS OF ANTIVIRAL AGENTS THAT INTERACT WITH THE CAPSID OF HUMAN RHINOVIRUSES | | Descriptor: | 5-(5-(6-CHLORO-4-(4,5-DIHYDRO-2-OXAZOLYL)PHENOXY)PENTYL)-3-METHYL ISOXAZOLE, HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP1), HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP2), ... | | Authors: | Badger, J, Smith, T.J, Rossmann, M.G. | | Deposit date: | 1988-10-03 | | Release date: | 1990-01-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural analysis of antiviral agents that interact with the capsid of human rhinoviruses.
Proteins, 6, 1989
|
|
2RS3
 
 | | STRUCTURAL ANALYSIS OF ANTIVIRAL AGENTS THAT INTERACT WITH THE CAPSID OF HUMAN RHINOVIRUSES | | Descriptor: | 5-(7-(5-HYDRO-4-ETHYL-2-OXAZOLYL)PHENOXY)HEPTYL)-3-METHYL ISOXAZOLE, HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP1), HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP2), ... | | Authors: | Badger, J, Smith, T.J, Rossmann, M.G. | | Deposit date: | 1988-10-03 | | Release date: | 1990-01-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural analysis of antiviral agents that interact with the capsid of human rhinoviruses.
Proteins, 6, 1989
|
|
2RS5
 
 | | STRUCTURAL ANALYSIS OF ANTIVIRAL AGENTS THAT INTERACT WITH THE CAPSID OF HUMAN RHINOVIRUSES | | Descriptor: | 5-(5-(4-(5-HYDRO-4-METHYL-2-OXAZOLYL)PHENOXY)PENTYL)-3-METHYL ISOXAZOLE, HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP1), HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP2), ... | | Authors: | Badger, J, Smith, T.J, Rossmann, M.G. | | Deposit date: | 1988-10-03 | | Release date: | 1990-01-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural analysis of antiviral agents that interact with the capsid of human rhinoviruses.
Proteins, 6, 1989
|
|
2RS1
 
 | | STRUCTURAL ANALYSIS OF ANTIVIRAL AGENTS THAT INTERACT WITH THE CAPSID OF HUMAN RHINOVIRUSES | | Descriptor: | 5-(7-(5-HYDRO-4-METHYL-2-OXAZOLYL)PHENOXY)HEPTYL)-3-METHYL ISOXAZOLE, HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP1), HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP2), ... | | Authors: | Badger, J, Smith, T.J, Rossmann, M.G. | | Deposit date: | 1988-10-03 | | Release date: | 1990-01-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural analysis of antiviral agents that interact with the capsid of human rhinoviruses.
Proteins, 6, 1989
|
|
