5L9W
 
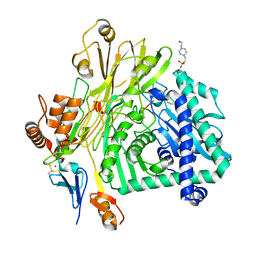 | | Crystal structure of the Apc core complex | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, Acetophenone carboxylase alpha subunit, ... | | Authors: | Warkentin, E, Weidenweber, S, Ermler, U. | | Deposit date: | 2016-06-11 | | Release date: | 2017-01-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the acetophenone carboxylase core complex: prototype of a new class of ATP-dependent carboxylases/hydrolases.
Sci Rep, 7, 2017
|
|
8ELZ
 
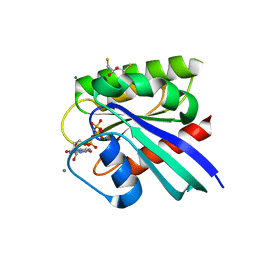 | | HRAS R97M Crystal Form 1 | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, CALCIUM ION, GTPase HRas, ... | | Authors: | Johnson, C.W, Mattos, C. | | Deposit date: | 2022-09-26 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.961 Å) | | Cite: | Allosteric site variants affect GTP hydrolysis on Ras.
Protein Sci., 32, 2023
|
|
5NKF
 
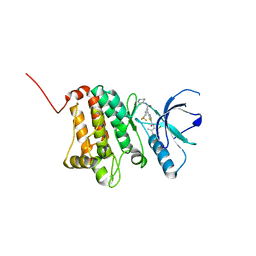 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 3b | | Descriptor: | Ephrin type-A receptor 2, ~{N}-(2-chloranyl-6-methyl-phenyl)-2-[[3-(piperidin-4-ylcarbamoyl)-5-(trifluoromethyl)phenyl]amino]-1,3-thiazole-5-carboxamide | | Authors: | Kudlinzki, D, Linhard, V.L, Witt, K, Gande, S.L, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2017-03-31 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.099 Å) | | Cite: | Chemoproteomics-Aided Medicinal Chemistry for the Discovery of EPHA2 Inhibitors.
ChemMedChem, 12, 2017
|
|
5K4U
 
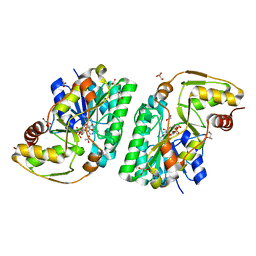 | | Three-dimensional structure of L-threonine 3-dehydrogenase from Trypanosoma brucei showing different active site loop conformations between dimer subunits, refined to 1.9 angstroms | | Descriptor: | ACETATE ION, GLYCEROL, L-threonine 3-dehydrogenase, ... | | Authors: | Adjogatse, E.K, Cooper, J.B, Erskine, P.T. | | Deposit date: | 2016-05-22 | | Release date: | 2017-11-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and function of L-threonine-3-dehydrogenase from the parasitic protozoan Trypanosoma brucei revealed by X-ray crystallography and geometric simulations.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6SBB
 
 | | Structure of type II terpene cyclase MstE from Scytonema (apo) | | Descriptor: | 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Moosmann, P, Ecker, F, Leopold-Messer, S, Cahn, J.K.B, Groll, M, Piel, J. | | Deposit date: | 2019-07-19 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A monodomain class II terpene cyclase assembles complex isoprenoid scaffolds.
Nat.Chem., 12, 2020
|
|
6DWR
 
 | | Trypsin serine protease modified with the protease inhibitor cyanobenzylsulfonylfluoride | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Cationic trypsin, ... | | Authors: | Luo, M, Eaton, C.N, Phillips-Piro, C.M. | | Deposit date: | 2018-06-27 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.319 Å) | | Cite: | Paired Spectroscopic and Crystallographic Studies of Protease Active Sites
Chemistryselect, 4, 2019
|
|
9Q7X
 
 | |
8CJB
 
 | | A268M variant of the CODH/ACS complex of C. hydrogenoformans | | Descriptor: | ACETATE ION, CO-methylating acetyl-CoA synthase, Carbon monoxide dehydrogenase, ... | | Authors: | Ruickoldt, J, Jeoung, J, Lennartz, F, Dobbek, H. | | Deposit date: | 2023-02-13 | | Release date: | 2024-02-21 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Coupling CO 2 Reduction and Acetyl-CoA Formation: The Role of a CO Capturing Tunnel in Enzymatic Catalysis.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
5LWO
 
 | | Structure of Spin-labelled T4 lysozyme mutant L115C-R119C-R1 at 100K | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Loll, B, Consentius, P, Gohlke, U, Mueller, R, Kaupp, M, Heinemann, U, Wahl, M.C, Risse, T. | | Deposit date: | 2016-09-18 | | Release date: | 2017-03-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.183 Å) | | Cite: | Internal Dynamics of the 3-Pyrroline-N-Oxide Ring in Spin-Labeled Proteins.
J Phys Chem Lett, 8, 2017
|
|
4RPD
 
 | |
8Y9Q
 
 | | b-glucosidase from Thermotoga profunda Tp-BGL | | Descriptor: | GLYCEROL, b-glucosidase | | Authors: | Guo, Y, Chen, A. | | Deposit date: | 2024-02-07 | | Release date: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural insights and functional characterization of a novel beta-glucosidase derived from Thermotoga profunda.
Biochem.Biophys.Res.Commun., 732, 2024
|
|
5NRQ
 
 | | Mtb TMK crystal structure in complex with compound 33 | | Descriptor: | 1-[1-[[5-(3-chloranylphenoxy)pyridin-3-yl]methyl]piperidin-4-yl]-5-methyl-pyrimidine-2,4-dione, CHLORIDE ION, Thymidylate kinase | | Authors: | Merceron, R, Song, L, Munier-Lehmann, H, Van Calenbergh, S, Savvides, S. | | Deposit date: | 2017-04-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mtb TMK crystal structure in complex with compound LS3112
To Be Published
|
|
9JJD
 
 | | Structural analysis of autophagy-related protein 8a in Drosophila melanogaster | | Descriptor: | Autophagy-related 8a, isoform A | | Authors: | Zhang, S.Q, Luo, X, Wu, D.X, Liu, J, Li, X.Y. | | Deposit date: | 2024-09-13 | | Release date: | 2025-03-26 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Crystal Structure of Autophagy-Associated Protein 8 at 1.36 angstrom Resolution and Its Inhibitory Interactions with Indole Analogs.
J.Agric.Food Chem., 73, 2025
|
|
5NKI
 
 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 4b | | Descriptor: | Ephrin type-A receptor 2, ~{N}-(2-chloranyl-6-methyl-phenyl)-2-[(3-methylsulfonyl-5-morpholin-4-yl-phenyl)amino]-1,3-thiazole-5-carboxamide | | Authors: | Kudlinzki, D, Linhard, V.L, Witt, K, Gande, S.L, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2017-03-31 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.675 Å) | | Cite: | Chemoproteomics-Aided Medicinal Chemistry for the Discovery of EPHA2 Inhibitors.
ChemMedChem, 12, 2017
|
|
5L10
 
 | | Crystal Structure of N-Acylhomoserine Lactone Dependent LuxR Family Transcriptionl Factor CepR2 from Burkholderia cenocepacia | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, GLYCEROL, ... | | Authors: | Kim, Y, Chhor, G, Jedrzejczak, R, Winan, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-07-28 | | Release date: | 2016-10-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structure of N-Acylhomoserine Lactone Dependent LuxR Family Transcriptionl Factor CepR2 from Burkholderia cenocepacia.
To Be Published
|
|
9G01
 
 | | Structure of carbon monoxide dehydrogenase/acetyl-CoA synthase (CODH/ACS) from Clostridium autoethanogenum (composite structure, closed and CO-bound state) | | Descriptor: | CARBON MONOXIDE, CO-methylating acetyl-CoA synthase, Carbon monoxide dehydrogenase/acetyl-CoA synthase beta subunit, ... | | Authors: | Yin, M.D, Lemaire, O.N, Wagner, T, Murphy, B.J. | | Deposit date: | 2024-07-06 | | Release date: | 2025-02-05 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Conformational dynamics of a multienzyme complex in anaerobic carbon fixation.
Science, 387, 2025
|
|
7ZBP
 
 | | Unspecific peroxygenase from Marasmius rotula | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Santillana, E, Romero, A. | | Deposit date: | 2022-03-24 | | Release date: | 2022-05-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural Characterization of Two Short Unspecific Peroxygenases: Two Different Dimeric Arrangements.
Antioxidants, 11, 2022
|
|
6SQL
 
 | | Crystal structure of M. tuberculosis InhA in complex with NAD+ and N-(3-(aminomethyl)phenyl)-5-chloro-3-methylbenzo[b]thiophene-2-sulfonamide | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ~{N}-[3-(aminomethyl)phenyl]-5-chloranyl-3-methyl-1-benzothiophene-2-sulfonamide | | Authors: | Mendes, V, Sabbah, M, Coyne, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2019-09-04 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Fragment-Based Design ofMycobacterium tuberculosisInhA Inhibitors.
J.Med.Chem., 63, 2020
|
|
6EPU
 
 | | The ATAD2 bromodomain in complex with compound 2 | | Descriptor: | (2~{S})-~{N}-(4-ethanoyl-1,3-thiazol-2-yl)piperazine-2-carboxamide, ATPase family AAA domain-containing protein 2, SULFATE ION | | Authors: | Sledz, P, Caflisch, A. | | Deposit date: | 2017-10-12 | | Release date: | 2018-10-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Hitting a Moving Target: Simulation and Crystallography Study of ATAD2 Bromodomain Blockers.
Acs Med.Chem.Lett., 11, 2020
|
|
5BJV
 
 | | X-ray structure of the PglF UDP-N-acetylglucosamine 4,6-dehydratase from Campylobacterjejuni, D396N/K397A variant in complex with UDP-N-acrtylglucosamine | | Descriptor: | 1,2-ETHANEDIOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, ... | | Authors: | Riegert, A.S, Thoden, J.B, Holden, H.M. | | Deposit date: | 2017-09-12 | | Release date: | 2017-11-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Biochemical Investigation of PglF from Campylobacter jejuni Reveals a New Mechanism for a Member of the Short Chain Dehydrogenase/Reductase Superfamily.
Biochemistry, 56, 2017
|
|
6IVC
 
 | | The full length of TGEV nsp1 | | Descriptor: | nsp1 protein | | Authors: | Shen, Z, Peng, G.Q. | | Deposit date: | 2018-12-03 | | Release date: | 2019-08-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A conserved region of nonstructural protein 1 from alphacoronaviruses inhibits host gene expression and is critical for viral virulence.
J.Biol.Chem., 294, 2019
|
|
9J24
 
 | |
6AQQ
 
 | | Crystal structure of Staphylococcus aureus biotin protein ligase in complex with inhibitor | | Descriptor: | (3aS,4S,6aR)-4-(5-{1-[(3-fluorophenyl)methyl]-1H-1,2,3-triazol-4-yl}pentyl)tetrahydro-1H-thieno[3,4-d]imidazol-2(3H)-one, Bifunctional ligase/repressor BirA | | Authors: | Cini, D.A, Wilce, M.C.J. | | Deposit date: | 2017-08-21 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Halogenation of Biotin Protein Ligase Inhibitors Improves Whole Cell Activity against Staphylococcus aureus.
ACS Infect Dis, 4, 2018
|
|
6ISA
 
 | | mCD226 | | Descriptor: | CD226 antigen | | Authors: | Wang, H, Qi, J, Zhang, S, Li, Y, Tan, S, Gao, G.F. | | Deposit date: | 2018-11-16 | | Release date: | 2018-12-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding mode of the side-by-side two-IgV molecule CD226/DNAM-1 to its ligand CD155/Necl-5.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5FIF
 
 | | Carboxyltransferase domain of a single-chain bacterial carboxylase | | Descriptor: | 1,2-ETHANEDIOL, Carboxylase | | Authors: | Hagmann, A, Hunkeler, M, Stuttfeld, E, Maier, T. | | Deposit date: | 2015-12-23 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.494 Å) | | Cite: | Hybrid Structure of a Dynamic Single-Chain Carboxylase from Deinococcus radiodurans.
Structure, 24, 2016
|
|
