6MZN
 
 | |
6MZU
 
 | |
6N0I
 
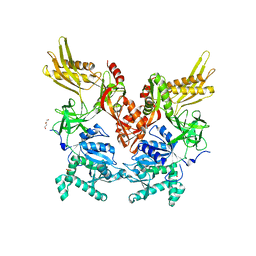 | | 2.60 Angstrom Resolution Crystal Structure of Elongation Factor G 2 from Pseudomonas putida. | | Descriptor: | DI(HYDROXYETHYL)ETHER, Elongation factor G 2, SULFATE ION | | Authors: | Minasov, G, Shuvalova, L, Wawrzak, Z, Cardona-Correa, A, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-11-07 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 2.60 Angstrom Resolution Crystal Structure of Elongation Factor G 2 from Pseudomonas putida.
To Be Published
|
|
6MRL
 
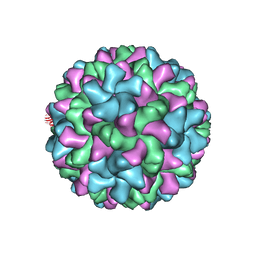 | | Cucumber Leaf Spot Virus | | Descriptor: | CALCIUM ION, p41 | | Authors: | Sherman, M.B, Smith, T.J. | | Deposit date: | 2018-10-14 | | Release date: | 2019-10-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Near-Atomic-Resolution Cryo-Electron Microscopy Structures of Cucumber Leaf Spot Virus and Red Clover Necrotic Mosaic Virus: Evolutionary Divergence at the Icosahedral Three-Fold Axes.
J.Virol., 94, 2020
|
|
4W4M
 
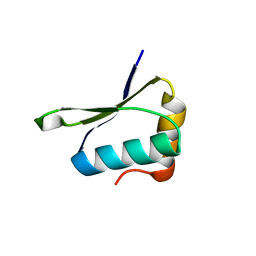 | | Crystal structure of PrgK 19-92 | | Descriptor: | Lipoprotein PrgK | | Authors: | Bergeron, J.R.C, Strynadka, N.C.J. | | Deposit date: | 2014-08-15 | | Release date: | 2014-10-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The Modular Structure of the Inner-Membrane Ring Component PrgK Facilitates Assembly of the Type III Secretion System Basal Body.
Structure, 23, 2015
|
|
4W4T
 
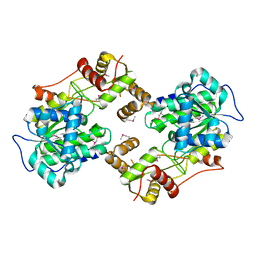 | | The crystal structure of the terminal R domain from the myxalamid PKS-NRPS biosynthetic pathway | | Descriptor: | ACETATE ION, MxaA | | Authors: | Tsai, S.C, Keasling, J.D, Luo, R, Barajas, J.F, Phelan, R.M, Schaub, A.J, Kliewer, J. | | Deposit date: | 2014-08-15 | | Release date: | 2015-08-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.845 Å) | | Cite: | Comprehensive Structural and Biochemical Analysis of the Terminal Myxalamid Reductase Domain for the Engineered Production of Primary Alcohols.
Chem.Biol., 22, 2015
|
|
6MSD
 
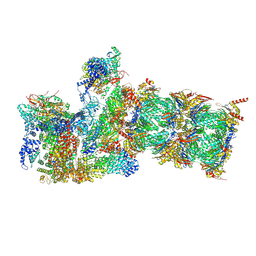 | | Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, ... | | Authors: | Mao, Y.D. | | Deposit date: | 2018-10-16 | | Release date: | 2018-12-05 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome.
Nature, 565, 2019
|
|
4W4X
 
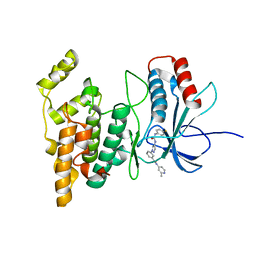 | | JNK2/3 in complex with 3-(4-{[(4-fluorophenyl)carbamoyl]amino}-1H-pyrazol-1-yl)-N-(2-methylpyridin-4-yl)benzamide | | Descriptor: | 3-(4-{[(4-fluorophenyl)carbamoyl]amino}-1H-pyrazol-1-yl)-N-(2-methylpyridin-4-yl)benzamide, c-jun NH2-terminal kinase 3 | | Authors: | Park, H, Iqbal, S, Hernandez, P, Mora, R, Zheng, K, Feng, Y, LoGrasso, P. | | Deposit date: | 2014-08-15 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural Basis and Biological Consequences for JNK2/3 Isoform Selective Aminopyrazoles.
Sci Rep, 5, 2015
|
|
6N13
 
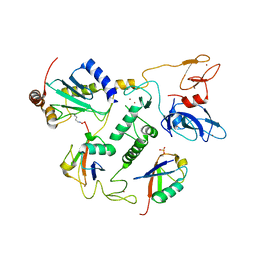 | | UbcH7-Ub Complex with R0RBR Parkin and phosphoubiquitin | | Descriptor: | E3 ubiquitin-protein ligase parkin, Ubiquitin-conjugating enzyme E2 L3, ZINC ION, ... | | Authors: | Condos, T.E.C, Dunkerley, K.M, Freeman, E.A, Barber, K.R, Aguirre, J.D, Chaugule, V.K, Xiao, Y, Konermann, L, Walden, H, Shaw, G.S. | | Deposit date: | 2018-11-08 | | Release date: | 2018-11-28 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Synergistic recruitment of UbcH7~Ub and phosphorylated Ubl domain triggers parkin activation.
EMBO J., 37, 2018
|
|
6N25
 
 | |
1E0O
 
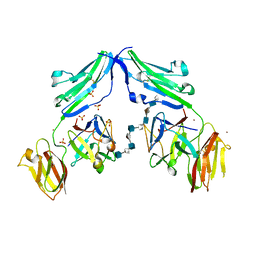 | | CRYSTAL STRUCTURE OF A TERNARY FGF1-FGFR2-HEPARIN COMPLEX | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, FIBROBLAST GROWTH FACTOR 1, FIBROBLAST GROWTH FACTOR RECEPTOR 2, ... | | Authors: | Pellegrini, L, Burke, D.F, von Delft, F, Mulloy, B, Blundell, T.L. | | Deposit date: | 2000-04-03 | | Release date: | 2000-10-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Fibroblast Growth Factor Receptor Ectodomain Bound to Ligand and Heparin
Nature, 407, 2000
|
|
6N2D
 
 | | Bacillus PS3 ATP synthase membrane region | | Descriptor: | ATP synthase subunit a, ATP synthase subunit b, ATP synthase subunit c | | Authors: | Guo, H, Rubinstein, J.L. | | Deposit date: | 2018-11-12 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of a bacterial ATP synthase.
Elife, 8, 2019
|
|
4W5C
 
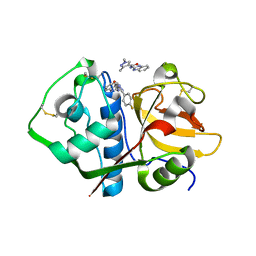 | | Crystal structure analysis of cruzain with three Fragments: 1 (N-(1H-benzimidazol-2-yl)-1,3-dimethyl-pyrazole-4-carboxamide), 6 (2-amino-4,6-difluorobenzothiazole) and 9 (N-(1H-benzimidazol-2-yl)-3-(4-fluorophenyl)-1H-pyrazole-4-carboxamide). | | Descriptor: | 4,6-difluoro-1,3-benzothiazol-2-amine, Cruzipain, N-(1H-benzimidazol-2-yl)-1,3-dimethyl-1H-pyrazole-4-carboxamide, ... | | Authors: | Tochowicz, A, McKerrow, J.H, Craik, C.S. | | Deposit date: | 2014-08-17 | | Release date: | 2015-04-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | Applying Fragments Based- Drug Design to identify multiple binding modes on cysteine protease.
To Be Published
|
|
6N2U
 
 | |
4W5Q
 
 | |
6N2Z
 
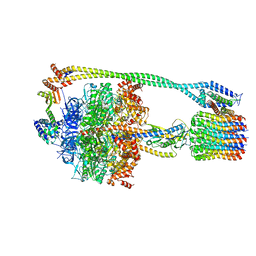 | | Bacillus PS3 ATP synthase class 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Guo, H, Rubinstein, J.L. | | Deposit date: | 2018-11-14 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of a bacterial ATP synthase.
Elife, 8, 2019
|
|
6MUH
 
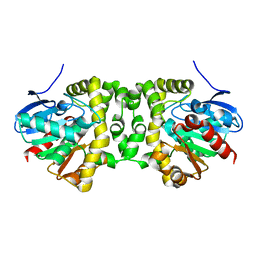 | | Fluoroacetate dehalogenase, room temperature structure solved by serial 1 degree oscillation crystallography | | Descriptor: | Fluoroacetate dehalogenase | | Authors: | Finke, A.D, Wierman, J.L, Pare-Labrosse, O, Sarrachini, A, Besaw, J, Mehrabi, P, Gruner, S.M, Miller, R.J.D. | | Deposit date: | 2018-10-23 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fixed-target serial oscillation crystallography at room temperature.
IUCrJ, 6, 2019
|
|
6MUO
 
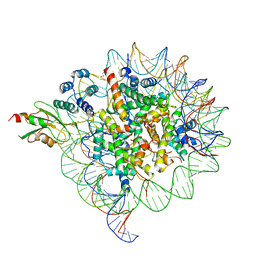 | |
1E4O
 
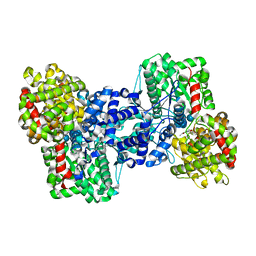 | | Phosphorylase recognition and phosphorolysis of its oligosaccharide substrate: answers to a long outstanding question | | Descriptor: | MALTODEXTRIN PHOSPHORYLASE, PYRIDOXAL-5'-PHOSPHATE, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Watson, K.A, McCleverty, C, Geremia, S, Cottaz, S, Driguez, H, Johnson, L.N. | | Deposit date: | 2000-07-11 | | Release date: | 2000-08-16 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Phosphorylase Recognition and Phosphorylysis of its Oligosaccharide Substrate: Answers to a Long Outstanding Question
Embo J., 18, 1999
|
|
6SED
 
 | | Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB in complex with galactose | | Descriptor: | ACETATE ION, Beta-galactosidase, FORMIC ACID, ... | | Authors: | Rutkiewicz, M, Bujacz, A, Kaminska, P, Bujacz, G. | | Deposit date: | 2019-07-29 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.233 Å) | | Cite: | Active Site Architecture and Reaction Mechanism Determination of Cold Adapted beta-d-galactosidase fromArthrobactersp. 32cB.
Int J Mol Sci, 20, 2019
|
|
6N5W
 
 | |
1DWJ
 
 | |
6N6N
 
 | | FtsY-NG high-resolution | | Descriptor: | ACETATE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Ataide, S.F, Faoro, C. | | Deposit date: | 2018-11-26 | | Release date: | 2019-10-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.877 Å) | | Cite: | Structural insights into the G-loop dynamics of E. coli FtsY NG domain.
J.Struct.Biol., 208, 2019
|
|
6N7E
 
 | |
6N09
 
 | |
