6SRY
 
 | | Kemp Eliminase HG3.17 mutant Q50S, E47N, N300D Complexed with Transition State Analog 6-Nitrobenzotriazole | | Descriptor: | 6-NITROBENZOTRIAZOLE, Kemp Eliminase HG3.17 Q50S, E47N,N300D, ... | | Authors: | Bloch, J.S, Pinkas, D.M, Hilvert, D. | | Deposit date: | 2019-09-06 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Contribution of Oxyanion Stabilization to Kemp Eliminase Efficiencyproficiency
Acs Catalysis, 2020
|
|
4LPW
 
 | |
3A1B
 
 | | Crystal structure of the DNMT3A ADD domain in complex with histone H3 | | Descriptor: | 1,2-ETHANEDIOL, DNA (cytosine-5)-methyltransferase 3A, Histone H3.1, ... | | Authors: | Otani, J, Arita, K, Ariyoshi, M, Shirakawa, M. | | Deposit date: | 2009-03-28 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.292 Å) | | Cite: | Structural basis for recognition of H3K4 methylation status by the DNA methyltransferase 3A ATRX-DNMT3-DNMT3L domain
Embo Rep., 10, 2009
|
|
5U5A
 
 | | Coiled Coil Peptide Metal Coordination Framework: Dimer Fold | | Descriptor: | COPPER (II) ION, Designed dimeric coiled coil peptide with two terpyridine side chains | | Authors: | Tavenor, N.A, Murnin, M.J, Horne, W.S. | | Deposit date: | 2016-12-06 | | Release date: | 2017-02-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | Supramolecular Metal-Coordination Polymers, Nets, and Frameworks from Synthetic Coiled-Coil Peptides.
J. Am. Chem. Soc., 139, 2017
|
|
5UHR
 
 | |
7QLF
 
 | |
5UFQ
 
 | | K-RasG12D(GNP)/R11.1.6 complex | | Descriptor: | CADMIUM ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Parker, J.A, Mattos, C. | | Deposit date: | 2017-01-05 | | Release date: | 2017-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | An engineered protein antagonist of K-Ras/B-Raf interaction.
Sci Rep, 7, 2017
|
|
4IX0
 
 | | Computational Design of an Unnatural Amino Acid Metalloprotein with Atomic Level Accuracy | | Descriptor: | NICKEL (II) ION, SULFATE ION, Unnatural Amino Acid Mediated Metalloprotein | | Authors: | Mills, J, Bolduc, J, Khare, S, Stoddard, B, Baker, D. | | Deposit date: | 2013-01-24 | | Release date: | 2013-08-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Computational design of an unnatural amino Acid dependent metalloprotein with atomic level accuracy.
J.Am.Chem.Soc., 135, 2013
|
|
4J8T
 
 | | Engineered Digoxigenin binder DIG10.2 | | Descriptor: | DIGOXIGENIN, Engineered Digoxigenin binder protein DIG10.2 | | Authors: | Stoddard, B.L, Doyle, L.A. | | Deposit date: | 2013-02-14 | | Release date: | 2013-06-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Computational design of ligand-binding proteins with high affinity and selectivity.
Nature, 501, 2013
|
|
7Q94
 
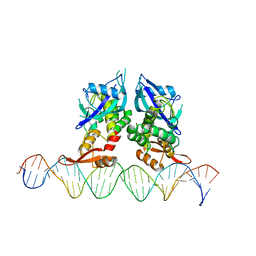 | | Crystal Structure of Agrobacterium tumefaciens NADQ, DNA complex. | | Descriptor: | DNA binding region (31-MER), NADQ transcription factor | | Authors: | Cianci, M, Minazzato, G, Heroux, A, Raffaelli, N, Sorci, L, Gasparrini, M. | | Deposit date: | 2021-11-11 | | Release date: | 2022-11-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Bacterial NadQ (COG4111) is a Nudix-like, ATP-responsive regulator of NAD biosynthesis.
J.Struct.Biol., 214, 2022
|
|
7Q91
 
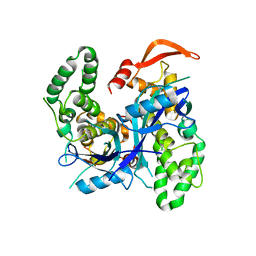 | | Crystal Structure of Agrobacterium tumefaciens NADQ, native form. | | Descriptor: | NADQ transcription factor, SODIUM ION | | Authors: | Cianci, M, Minazzato, G, Heroux, A, Raffaelli, N, Sorci, L, Gasparrini, M. | | Deposit date: | 2021-11-11 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Bacterial NadQ (COG4111) is a Nudix-like, ATP-responsive regulator of NAD biosynthesis.
J.Struct.Biol., 214, 2022
|
|
7Q93
 
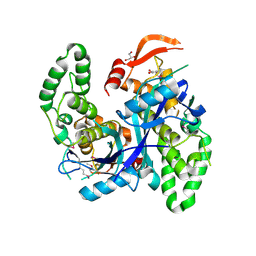 | | Crystal Structure of Agrobacterium tumefaciens NADQ, NAD complex. | | Descriptor: | GLYCEROL, NADQ transcription factor, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Cianci, M, Minazzato, G, Heroux, A, Raffaelli, N, Sorci, L, Gasparrini, M. | | Deposit date: | 2021-11-11 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Bacterial NadQ (COG4111) is a Nudix-like, ATP-responsive regulator of NAD biosynthesis.
J.Struct.Biol., 214, 2022
|
|
7Q92
 
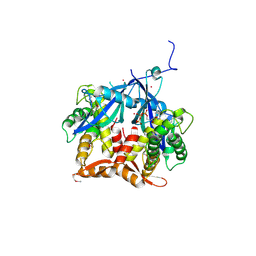 | | Crystal Structure of Agrobacterium tumefaciens NADQ, ATP complex. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, NADQ transcription factor, POTASSIUM ION, ... | | Authors: | Cianci, M, Minazzato, G, Heroux, A, Raffaelli, N, Sorci, L, Gasparrini, M. | | Deposit date: | 2021-11-11 | | Release date: | 2022-11-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Bacterial NadQ (COG4111) is a Nudix-like, ATP-responsive regulator of NAD biosynthesis.
J.Struct.Biol., 214, 2022
|
|
4LNY
 
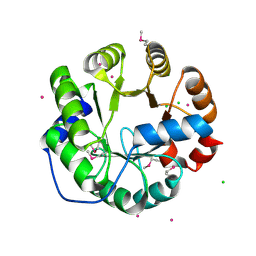 | | Crystal Structure of Engineered Protein, Northeast Structural Genomics Consortium Target OR422 | | Descriptor: | CADMIUM ION, CHLORIDE ION, Engineered Protein OR422 | | Authors: | Vorobiev, S, Su, M, Bjelic, S, Kipnis, Y, Wang, L, Sahdev, S, Xiao, R, Maglaqui, M, Kogan, S, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-07-12 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.929 Å) | | Cite: | Crystal Structure of Engineered Protein OR422.
To be Published
|
|
7QWC
 
 | |
5T56
 
 | |
7QWE
 
 | |
7JQU
 
 | |
7JQS
 
 | |
7JQR
 
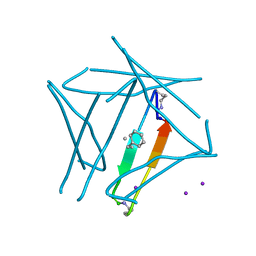 | | Abeta 16-36 beta-hairpin mimic with E22G Arctic mutation | | Descriptor: | Abeta 16-36 beta-hairpin mimic VAL-ORN-LYS-LEU-VAL-MEA-PHE-ALA-GLY-ORN-ALA-ILE-ILE-GLY-LEU-MET, IODIDE ION | | Authors: | Kreutzer, A.G, McKnelly, K.J, Nowick, J.S. | | Deposit date: | 2020-08-11 | | Release date: | 2021-09-08 | | Last modified: | 2022-03-30 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Effects of Familial Alzheimer's Disease Mutations on the Assembly of a beta-Hairpin Peptide Derived from A beta 16-36 .
Biochemistry, 61, 2022
|
|
7QWD
 
 | |
7QWB
 
 | |
7JXN
 
 | |
7QWA
 
 | |
5V64
 
 | |
