5K8J
 
 | | Structure of Caulobacter crescentus VapBC1 (apo form) | | Descriptor: | GLYCEROL, Ribonuclease VapC, VapB family protein | | Authors: | Bendtsen, K.L, Xu, K, Luckmann, M, Brodersen, D.E. | | Deposit date: | 2016-05-30 | | Release date: | 2016-12-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Toxin inhibition in C. crescentus VapBC1 is mediated by a flexible pseudo-palindromic protein motif and modulated by DNA binding.
Nucleic Acids Res., 45, 2017
|
|
5D4U
 
 | | SAM-bound HcgC from Methanocaldococcus jannaschii | | Descriptor: | S-ADENOSYLMETHIONINE, SULFATE ION, Uncharacterized protein MJ0489 | | Authors: | Fujishiro, T, Ermler, U, Shima, S. | | Deposit date: | 2015-08-09 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of HcgC as a SAM-Dependent Pyridinol Methyltransferase in [Fe]-Hydrogenase Cofactor Biosynthesis.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5D5Q
 
 | | HcgB from Methanocaldococcus jannaschii with the pyridinol derived from FeGP cofactor of [Fe]-hydrogenase | | Descriptor: | (4,6-dihydroxy-3,5-dimethylpyridin-2-yl)acetic acid, Uncharacterized protein MJ0488, Guanylyltransferase | | Authors: | Fujishiro, T, Ermler, U, Shima, S. | | Deposit date: | 2015-08-11 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Towards artificial methanogenesis: biosynthesis of the [Fe]-hydrogenase cofactor and characterization of the semi-synthetic hydrogenase.
Faraday Discuss., 198, 2017
|
|
5L6L
 
 | | Structure of Caulobacter crescentus VapBC1 bound to operator DNA | | Descriptor: | DNA (27-MER), Ribonuclease VapC, VapB family protein | | Authors: | Bendtsen, K.L, Xu, K, Luckmann, M, Brodersen, D.E. | | Deposit date: | 2016-05-30 | | Release date: | 2016-12-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Toxin inhibition in C. crescentus VapBC1 is mediated by a flexible pseudo-palindromic protein motif and modulated by DNA binding.
Nucleic Acids Res., 45, 2017
|
|
5LHP
 
 | | The p-aminobenzamidine active site inhibited catalytic domain of murine urokinase-type plasminogen activator in complex with the allosteric inhibitory nanobody Nb7 | | Descriptor: | 1,2-ETHANEDIOL, Camelid-Derived Antibody Fragment, P-AMINO BENZAMIDINE, ... | | Authors: | Kromann-Hansen, T, Lange, E.L, Sorensen, H.P, Ghassabeh, G.H, Huang, M, Jensen, J.K, Muyldermans, S, Declerck, P.J, Andreasen, P.A. | | Deposit date: | 2016-07-12 | | Release date: | 2017-06-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Discovery of a novel conformational equilibrium in urokinase-type plasminogen activator.
Sci Rep, 7, 2017
|
|
5LHN
 
 | | The catalytic domain of murine urokinase-type plasminogen activator in complex with the allosteric inhibitory nanobody Nb7 | | Descriptor: | 1,2-ETHANEDIOL, Camelid-Derived Antibody Fragment Nb7, SULFATE ION, ... | | Authors: | Kromann-Hansen, T, Lange, E.L, Sorensen, H.P, Ghassabeh, G.H, Huang, M, Jensen, J.K, Muyldermans, S, Declerck, P, Andreasen, P.A. | | Deposit date: | 2016-07-12 | | Release date: | 2017-06-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery of a novel conformational equilibrium in urokinase-type plasminogen activator.
Sci Rep, 7, 2017
|
|
5LDJ
 
 | |
5LHS
 
 | | The ligand free catalytic domain of murine urokinase-type plasminogen activator | | Descriptor: | NICKEL (II) ION, SULFATE ION, Urokinase-type plasminogen activator | | Authors: | Kromann-Hansen, T, Lange, E.L, Sorensen, H.P, Ghassabeh, G.H, Huang, M, Jensen, J.K, Muyldermans, S, Declerck, P.J, Andreasen, P.A. | | Deposit date: | 2016-07-12 | | Release date: | 2017-06-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.047 Å) | | Cite: | Discovery of a novel conformational equilibrium in urokinase-type plasminogen activator.
Sci Rep, 7, 2017
|
|
5LDK
 
 | |
5LDP
 
 | |
5LSV
 
 | | X-ray crystal structure of AA13 LPMO | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, AoAA13, ZINC ION, ... | | Authors: | Frandsen, K.E.H, Poulsen, J.-C.N, Tovborg, M, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2016-09-05 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Learning from oligosaccharide soaks of crystals of an AA13 lytic polysaccharide monooxygenase: crystal packing, ligand binding and active-site disorder.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5M2V
 
 | | Structure of GluK1 ligand-binding domain (S1S2) in complex with (2S,4R)-4-(2-carboxyphenoxy)pyrrolidine-2-carboxylic acid at 3.18 A resolution | | Descriptor: | (2~{S},4~{R})-4-(2-carboxyphenoxy)pyrrolidine-2-carboxylic acid, CHLORIDE ION, Glutamate receptor ionotropic, ... | | Authors: | Frydenvang, K, Kastrup, J.S, Kristensen, C.M. | | Deposit date: | 2016-10-13 | | Release date: | 2017-01-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Design and Synthesis of a Series of l-trans-4-Substituted Prolines as Selective Antagonists for the Ionotropic Glutamate Receptors Including Functional and X-ray Crystallographic Studies of New Subtype Selective Kainic Acid Receptor Subtype 1 (GluK1) Antagonist (2S,4R)-4-(2-Carboxyphenoxy)pyrrolidine-2-carboxylic Acid.
J. Med. Chem., 60, 2017
|
|
6NUI
 
 | | Human Guanylate Kinase | | Descriptor: | Guanylate kinase | | Authors: | Sabo, T.M, Khan, N, Ban, D, Trigo-Mourino, P, Carneiro, M.G, Trent, J.O, Konrad, M, Lee, D. | | Deposit date: | 2019-02-01 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure and functional investigation of human guanylate kinase reveals allosteric networking and a crucial role for the enzyme in cancer.
J.Biol.Chem., 294, 2019
|
|
8ONI
 
 | | Human insulin in complex with the analytical antibody S1 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of analytical antibody S1 Fab, Insulin A chain, ... | | Authors: | Johansson, E. | | Deposit date: | 2023-04-03 | | Release date: | 2024-04-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Macromolecular structure of insulin and IgE clonality impacts IgE-mediated activation of sensitized basophils
To Be Published
|
|
5LDO
 
 | | Crystal structure of E.coli LigT complexed with 3'-AMP | | Descriptor: | RNA 2',3'-cyclic phosphodiesterase, [(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-hydroxy-2-(hydroxymethyl)oxolan-3-yl] dihydrogen phosphate | | Authors: | Myllykoski, M, Kursula, P. | | Deposit date: | 2016-06-27 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.752 Å) | | Cite: | Structural aspects of nucleotide ligand binding by a bacterial 2H phosphoesterase.
PLoS ONE, 12, 2017
|
|
5MIQ
 
 | | Crystal Structure of Lactococcus lactis Thioredoxin Reductase Exposed to Visible Light (60 min) | | Descriptor: | DI(HYDROXYETHYL)ETHER, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Skjoldager, N, Bang, M.B, Svensson, B, Hagglund, P, Harris, P. | | Deposit date: | 2016-11-29 | | Release date: | 2017-04-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The structure of Lactococcus lactis thioredoxin reductase reveals molecular features of photo-oxidative damage.
Sci Rep, 7, 2017
|
|
5LHQ
 
 | | The EGR-cmk active site inhibited catalytic domain of murine urokinase-type plasminogen activator in complex with the allosteric inhibitory nanobody Nb7 | | Descriptor: | 1,2-ETHANEDIOL, Camelid-Derived Antibody Fragment Nb7, L-alpha-glutamyl-N-{(1S)-4-{[amino(iminio)methyl]amino}-1-[(1S)-2-chloro-1-hydroxyethyl]butyl}glycinamide, ... | | Authors: | Kromann-Hansen, T, Lange, E.L, Sorensen, H.P, Ghassabeh, G.H, Huang, M, Jensen, J.K, Muyldermans, S, Declerck, P.J, Andreasen, P.A. | | Deposit date: | 2016-07-12 | | Release date: | 2017-06-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of a novel conformational equilibrium in urokinase-type plasminogen activator.
Sci Rep, 7, 2017
|
|
5LS2
 
 | | Receptor mediated chitin perception in legumes is functionally seperable from Nod factor perception | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LysM type receptor kinase, SULFATE ION | | Authors: | Bozsoki, Z, Cheng, J, Feng, F, Gysel, K, Andersen, K.R, Oldroyd, G, Blaise, M, Radutoiu, S, Stougaard, J. | | Deposit date: | 2016-08-22 | | Release date: | 2017-08-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Receptor-mediated chitin perception in legume roots is functionally separable from Nod factor perception.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5MJK
 
 | | Crystal Structure of Lactococcus lactis Thioredoxin Reductase (FO conformation) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, Thioredoxin reductase | | Authors: | Skjoldager, N, Bang, M.B, Svensson, B, Hagglund, P, Harris, P. | | Deposit date: | 2016-12-01 | | Release date: | 2017-04-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of Lactococcus lactis thioredoxin reductase reveals molecular features of photo-oxidative damage.
Sci Rep, 7, 2017
|
|
5MNT
 
 | | Bacteriophage Qbeta maturation protein | | Descriptor: | A2 maturation protein | | Authors: | Rumnieks, J, Tars, K. | | Deposit date: | 2016-12-13 | | Release date: | 2017-01-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Crystal Structure of the Maturation Protein from Bacteriophage Q beta.
J. Mol. Biol., 429, 2017
|
|
5M8B
 
 | |
5LDM
 
 | |
5LFN
 
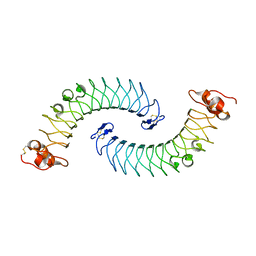 | | Crystal structure of human chondroadherin | | Descriptor: | CHLORIDE ION, Chondroadherin | | Authors: | Ramisch, S, Pramhed, A, Logan, D.T. | | Deposit date: | 2016-07-03 | | Release date: | 2016-12-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human chondroadherin: solving a difficult molecular-replacement problem using de novo models.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
1RKI
 
 | | Structure of pag5_736 from P. aerophilum with three disulphide bonds | | Descriptor: | ACETATE ION, CHLORIDE ION, HEXAETHYLENE GLYCOL, ... | | Authors: | Beeby, M, Ryttersgaard, C, Boutz, D.R, Perry, L.J, Yeates, T.O. | | Deposit date: | 2003-11-21 | | Release date: | 2005-01-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Genomics of Disulfide Bonding and Protein Stabilization in Thermophiles.
Plos Biol., 3, 2005
|
|
5MFW
 
 | | Crystal structure of the GluK1 ligand-binding domain in complex with kainate and BPAM-121 at 2.10 A resolution | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, 7-chloro-4-(2-fluoroethyl)-2,3-dihydro-1,2,4-benzothiadiazine 1,1-dioxide, ACETATE ION, ... | | Authors: | Larsen, A.P, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2016-11-18 | | Release date: | 2017-04-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification and Structure-Function Study of Positive Allosteric Modulators of Kainate Receptors.
Mol. Pharmacol., 91, 2017
|
|
