2AH9
 
 | | Crystal Structure of Human M340H-Beta-1,4-Galactosyltransferase-I (M340H-B4Gal-T1) in Complex with Chitotriose | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ramasamy, V, Ramakrishnan, B, Boeggeman, E, Ratner, D.M, Seeberger, P.H, Qasba, P.K. | | Deposit date: | 2005-07-27 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Oligosaccharide Preferences of beta1,4-Galactosyltransferase-I: Crystal Structures of Met340His Mutant of Human beta1,4-Galactosyltransferase-I with a Pentasaccharide and Trisaccharides of the N-Glycan Moiety
J.Mol.Biol., 353, 2005
|
|
4C1B
 
 | |
3L2Y
 
 | | The structure of C-reactive protein bound to phosphoethanolamine | | Descriptor: | C-reactive protein, CALCIUM ION, PHOSPHORIC ACID MONO-(2-AMINO-ETHYL) ESTER | | Authors: | Mikolajek, H, Kolstoe, S.E, Wood, S.P, Pepys, M.B. | | Deposit date: | 2009-12-15 | | Release date: | 2010-12-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of ligand specificity in the human pentraxins, C-reactive protein and serum amyloid P component.
J.Mol.Recognit., 24, 2011
|
|
3ZOW
 
 | | Crystal Structure of Wild Type Nitrosomonas europaea Cytochrome c552 | | Descriptor: | CYTOCHROME C-552, HEME C | | Authors: | Hersleth, H.-P, Can, M, Krucinska, J, Zoppellaro, G, Andersen, N.H, Karlsen, S, Wedekind, J.E, Andersson, K.K, Bren, K.L. | | Deposit date: | 2013-02-25 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Characterization of Nitrosomonas Europaea Cytochrome C-552 Variants with Marked Differences in Electronic Structure.
Chembiochem, 14, 2013
|
|
3KTW
 
 | | Crystal structure of the SRP19/S-domain SRP RNA complex of Sulfolobus solfataricus | | Descriptor: | MAGNESIUM ION, POTASSIUM ION, SRP RNA, ... | | Authors: | Wild, K, Bange, G, Bozkurt, G, Sinning, I. | | Deposit date: | 2009-11-26 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into the assembly of the human and archaeal signal recognition particles.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3L1T
 
 | | E. coli NrfA sulfite ocmplex | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Cytochrome c-552, ... | | Authors: | Clarke, T.A, Hemmings, A.M, Butt, J.N. | | Deposit date: | 2009-12-14 | | Release date: | 2010-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Kinetic and thermodynamic resolution of the interactions between sulfite and the pentahaem cytochrome NrfA from Escherichia coli
Biochem.J., 431, 2010
|
|
3ZKB
 
 | | CRYSTAL STRUCTURE OF THE ATPASE REGION OF Mycobacterium tuberculosis GyrB WITH AMPPNP | | Descriptor: | DNA GYRASE SUBUNIT B, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Agrawal, A, Roue, M, Spitzfaden, C, Petrella, S, Aubry, A, Volker, C, Mossakowska, D, Hann, M, Bax, B, Mayer, C. | | Deposit date: | 2013-01-22 | | Release date: | 2013-09-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mycobacterium Tuberculosis DNA Gyrase ATPase Domain Structures Suggest a Dissociative Mechanism that Explains How ATP Hydrolysis is Coupled to Domain Motion.
Biochem.J., 456, 2013
|
|
3KQX
 
 | | Structure of a protease 1 | | Descriptor: | CARBONATE ION, M17 leucyl aminopeptidase, NONAETHYLENE GLYCOL, ... | | Authors: | McGowan, S, Whisstock, J.C. | | Deposit date: | 2009-11-17 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure of the Plasmodium falciparum M17 aminopeptidase and significance for the design of drugs targeting the neutral exopeptidases
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
483D
 
 | |
3MG6
 
 | | Structure of yeast 20S open-gate proteasome with Compound 6 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, N~2~-{[(1S)-6-methoxy-3-oxo-2,3-dihydro-1H-inden-1-yl]acetyl}-N-{(1S)-1-[(4-methylbenzyl)carbamoyl]-3-phenylpropyl}-L-threoninamide, ... | | Authors: | Sintchak, M.D. | | Deposit date: | 2010-04-05 | | Release date: | 2011-05-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Characterization of a new series of non-covalent proteasome inhibitors with exquisite potency and selectivity for the 20S beta5-subunit.
Biochem.J., 430, 2010
|
|
4DCX
 
 | | X-ray structure of NikA in complex with Fe(1R,2R)-N,N'-Bis(2-pyridylmethyl)-N,N'-dicarboxymethyl-1,2-cyclohexanediamine | | Descriptor: | ACETATE ION, GLYCEROL, Nickel-binding periplasmic protein, ... | | Authors: | Cherrier, M.V, Girgenti, E, Amara, P, Iannello, M, Marchi-Delapierre, C, Fontecilla-Camps, J.C, Menage, S, Cavazza, C. | | Deposit date: | 2012-01-18 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of the periplasmic nickel-binding protein NikA provides insights for artificial metalloenzyme design.
J.Biol.Inorg.Chem., 17, 2012
|
|
4DDF
 
 | | Computationally Designed Self-assembling Octahedral Cage protein, O333, Crystallized in space group P4 | | Descriptor: | CHLORIDE ION, Propanediol utilization polyhedral body protein PduT, SULFATE ION | | Authors: | Sawaya, M.R, King, N.P, Sheffler, W, Baker, D, Yeates, T.O. | | Deposit date: | 2012-01-18 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Computational design of self-assembling protein nanomaterials with atomic level accuracy.
Science, 336, 2012
|
|
4DQ7
 
 | |
4DR3
 
 | | Crystal structure of the Thermus thermophilus (HB8) 30S ribosomal subunit with streptomycin bound | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Demirci, H, Murphy IV, F, Murphy, E, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2012-02-16 | | Release date: | 2012-11-14 | | Last modified: | 2013-01-30 | | Method: | X-RAY DIFFRACTION (3.348 Å) | | Cite: | A structural basis for streptomycin-induced misreading of the genetic code.
Nat Commun, 4, 2013
|
|
3MEN
 
 | |
4DV5
 
 | | Crystal structure of the Thermus thermophilus 30S ribosomal subunit with a 16S rRNA mutation, A914G, bound with streptomycin | | Descriptor: | 16S rRNA, MAGNESIUM ION, STREPTOMYCIN, ... | | Authors: | Demirci, H, Murphy IV, F, Murphy, E, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2012-02-22 | | Release date: | 2013-02-27 | | Method: | X-RAY DIFFRACTION (3.683 Å) | | Cite: | A structural basis for streptomycin resistance
To be Published
|
|
4DM5
 
 | |
3MG0
 
 | | Structure of yeast 20S proteasome with bortezomib | | Descriptor: | N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE, Proteasome component C1, Proteasome component C11, ... | | Authors: | Sintchak, M.D. | | Deposit date: | 2010-04-05 | | Release date: | 2011-05-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Characterization of a new series of non-covalent proteasome inhibitors with exquisite potency and selectivity for the 20S beta5-subunit.
Biochem.J., 430, 2010
|
|
3MHI
 
 | | Crystal structure of human carbonic anhydrase isozyme II with 4-{[(5-nitro-6-oxo-1,6-dihydro-4-pyrimidinyl)amino]methyl}benzenesulfonamide | | Descriptor: | 4-{[(5-nitro-6-oxo-1,6-dihydropyrimidin-4-yl)amino]methyl}benzenesulfonamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Grazulis, S, Manakova, E, Golovenko, D. | | Deposit date: | 2010-04-08 | | Release date: | 2010-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 4-[N-(Substituted 4-pyrimidinyl)amino]benzenesulfonamides as inhibitors of carbonic anhydrase isozymes I, II, VII, and XIII
Bioorg.Med.Chem., 18, 2010
|
|
3MK7
 
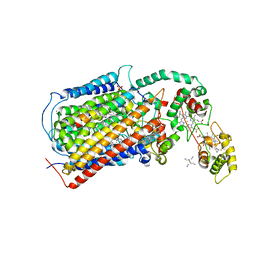 | | The structure of CBB3 cytochrome oxidase | | Descriptor: | 30-mer peptide, CALCIUM ION, COPPER (II) ION, ... | | Authors: | Buschmann, S, Warkentin, E, Michel, H, Ermler, U. | | Deposit date: | 2010-04-14 | | Release date: | 2010-08-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The Structure of cbb3 Cytochrome Oxidase Provides Insights into Proton Pumping
Science, 329, 2010
|
|
4C0Z
 
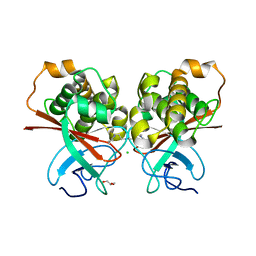 | | The N-terminal domain of the Streptococcus pyogenes pilus tip adhesin Cpa | | Descriptor: | ANCILLARY PROTEIN 2, CHLORIDE ION, GLYCEROL, ... | | Authors: | Linke-Winnebeck, C, Paterson, N, Baker, E.N. | | Deposit date: | 2013-08-08 | | Release date: | 2013-11-20 | | Last modified: | 2014-01-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Model for the Covalent Adhesion of the Streptococcus Pyogenes Pilus Through a Thioester Bond.
J.Biol.Chem., 289, 2014
|
|
4CE4
 
 | | 39S large subunit of the porcine mitochondrial ribosome | | Descriptor: | 16S Ribosomal RNA, ICT1, MRPL13, ... | | Authors: | Greber, B.J, Boehringer, D, Leitner, A, Bieri, P, Voigts-Hoffmann, F, Erzberger, J.P, Leibundgut, M, Aebersold, R, Ban, N. | | Deposit date: | 2013-11-08 | | Release date: | 2013-12-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Architecture of the Large Subunit of the Mammalian Mitochondrial Ribosome.
Nature, 505, 2014
|
|
3NEY
 
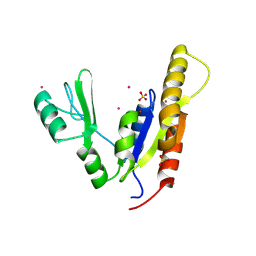 | | Crystal structure of the kinase domain of MPP1/p55 | | Descriptor: | 55 kDa erythrocyte membrane protein, SULFATE ION, UNKNOWN ATOM OR ION | | Authors: | Shen, Y, Tong, Y, Zhong, N, Guan, X, Tempel, W, MacKenzie, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-06-09 | | Release date: | 2010-08-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structure of the kinase domain of MPP1/p55
To be Published
|
|
3NG9
 
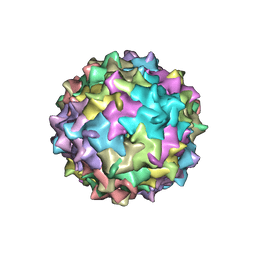 | | Structure to Function Correlations for Adeno-associated Virus Serotype 1 | | Descriptor: | 6-AMINOPYRIMIDIN-2(1H)-ONE, ADENINE, Capsid protein | | Authors: | Govindasamy, L, Miller, E.B, Gurda, B, McKenna, R, Zolotukhin, S, Muzyczka, N, Agbandje-McKenna, M. | | Deposit date: | 2010-06-11 | | Release date: | 2011-12-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure to Function Correlations for Adeno-Associated Virus serotype 1
To be Published
|
|
3N7A
 
 | | Crystal structure of 3-dehydroquinate dehydratase from Mycobacterium tuberculosis in complex with inhibitor 2 | | Descriptor: | 2,3 -ANHYDRO-QUINIC ACID, 3-dehydroquinate dehydratase, GLYCEROL | | Authors: | Dias, M.V.B, Snee, W.C, Bromfield, K.M, Payne, R, Palaninathan, S.K, Ciulli, A, Howard, N.I, Abell, C, Sacchettini, J.C, Blundell, T.L. | | Deposit date: | 2010-05-26 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural investigation of inhibitor designs targeting 3-dehydroquinate dehydratase from the shikimate pathway of Mycobacterium tuberculosis.
Biochem.J., 436, 2011
|
|
