4V4V
 
 | |
5IRT
 
 | |
1KUG
 
 | | Crystal Structure of a Taiwan Habu Venom Metalloproteinase complexed with its endogenous inhibitor pENW | | Descriptor: | CADMIUM ION, ENW, metalloproteinase | | Authors: | Huang, K.F, Chiou, S.H, Ko, T.P, Wang, A.H.J. | | Deposit date: | 2002-01-22 | | Release date: | 2002-07-10 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Determinants of the inhibition of a Taiwan habu venom metalloproteinase by its endogenous inhibitors revealed by X-ray crystallography and synthetic inhibitor analogues.
Eur.J.Biochem., 269, 2002
|
|
1KUK
 
 | | Crystal Structure of a Taiwan Habu Venom Metalloproteinase complexed with pEKW. | | Descriptor: | CADMIUM ION, EKW, metalloproteinase | | Authors: | Huang, K.F, Chiou, S.H, Ko, T.P, Wang, A.H.J. | | Deposit date: | 2002-01-22 | | Release date: | 2002-07-10 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Determinants of the inhibition of a Taiwan habu venom metalloproteinase by its endogenous inhibitors revealed by X-ray crystallography and synthetic inhibitor analogues.
Eur.J.Biochem., 269, 2002
|
|
1KUI
 
 | | Crystal Structure of a Taiwan Habu Venom Metalloproteinase complexed with pEQW. | | Descriptor: | CADMIUM ION, EQW, metalloproteinase | | Authors: | Huang, K.F, Chiou, S.H, Ko, T.P, Wang, A.H.J. | | Deposit date: | 2002-01-22 | | Release date: | 2002-07-10 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Determinants of the inhibition of a Taiwan habu venom metalloproteinase by its endogenous inhibitors revealed by X-ray crystallography and synthetic inhibitor analogues.
Eur.J.Biochem., 269, 2002
|
|
8IHI
 
 | | Cryo-EM structure of HCA2-Gi complex with acifran | | Descriptor: | (5~{S})-5-methyl-4-oxidanylidene-5-phenyl-furan-2-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Suzuki, S, Nishikawa, K, Suzuki, H, Fujiyoshi, Y. | | Deposit date: | 2023-02-22 | | Release date: | 2023-08-30 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Structural basis of hydroxycarboxylic acid receptor signaling mechanisms through ligand binding.
Nat Commun, 14, 2023
|
|
8IHF
 
 | | Cryo-EM structure of HCA2-Gi complex with MK6892 | | Descriptor: | 2-[[2,2-dimethyl-3-[3-(5-oxidanylpyridin-2-yl)-1,2,4-oxadiazol-5-yl]propanoyl]amino]cyclohexene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Suzuki, S, Nishikawa, K, Suzuki, H, Fujiyoshi, Y. | | Deposit date: | 2023-02-22 | | Release date: | 2023-08-30 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structural basis of hydroxycarboxylic acid receptor signaling mechanisms through ligand binding.
Nat Commun, 14, 2023
|
|
8IHJ
 
 | | Cryo-EM structure of HCA3-Gi complex with acifran | | Descriptor: | (5~{S})-5-methyl-4-oxidanylidene-5-phenyl-furan-2-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Suzuki, S, Nishikawa, K, Suzuki, H, Fujiyoshi, Y. | | Deposit date: | 2023-02-22 | | Release date: | 2023-08-30 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural basis of hydroxycarboxylic acid receptor signaling mechanisms through ligand binding.
Nat Commun, 14, 2023
|
|
8I95
 
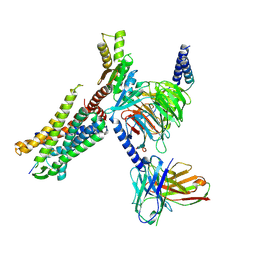 | | Structure of EP54-C3aR-Go complex | | Descriptor: | Antibody fragment - ScFv16, C3a anaphylatoxin chemotactic receptor, EP54 ligand, ... | | Authors: | Yadav, M.K, Yadav, R, Maharana, J, Sarma, P, Banerjee, R, Shukla, A.K, Gati, C. | | Deposit date: | 2023-02-06 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Molecular basis of anaphylatoxin binding, activation, and signaling bias at complement receptors.
Cell, 186, 2023
|
|
8IA2
 
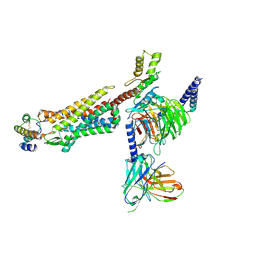 | | Structure of C5a bound human C5aR1 in complex with Go (Composite map) | | Descriptor: | Antibody fragment - ScFv16, C5a anaphylatoxin, C5a anaphylatoxin chemotactic receptor 1, ... | | Authors: | Yadav, M.K, Yadav, R, Maharana, J, Banerjee, R, Shukla, A.K, Gati, C. | | Deposit date: | 2023-02-07 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Molecular basis of anaphylatoxin binding, activation, and signaling bias at complement receptors.
Cell, 186, 2023
|
|
8IA7
 
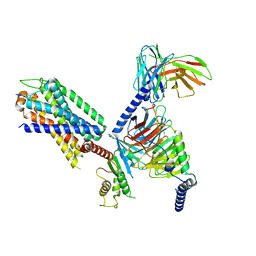 | | Structural insights into human brain gut peptide cholecystokinin receptors | | Descriptor: | CCK-8, Gastrin/cholecystokinin type B receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Ding, Y, Zhang, H, Liao, Y, Chen, L, Ji, S. | | Deposit date: | 2023-02-08 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into human brain-gut peptide cholecystokinin receptors.
Cell Discov, 8, 2022
|
|
8IM7
 
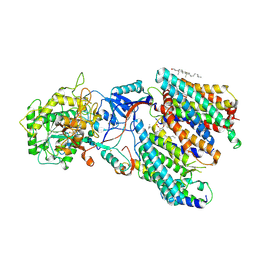 | | Human gamma-secretase treated with ganglioside GM1 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhou, R, Yang, G, Shi, Y. | | Deposit date: | 2023-03-06 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Preferential Regulation of Gamma-Secretase-Mediated Cleavage of APP by Ganglioside GM1 Reveals a Potential Therapeutic Target for Alzheimer's Disease.
Adv Sci, 10, 2023
|
|
6NCR
 
 | |
8DAY
 
 | | Crystal Structure of DMATS1 prenyltransferase in complex with L-Tyr and DMSPP | | Descriptor: | DIMETHYLALLYL S-THIOLODIPHOSPHATE, Dimethylallyltryptophan synthase 1, TYROSINE | | Authors: | Eaton, S.A, Ronnebaum, T.A, Roose, B.W, Christianson, D.W. | | Deposit date: | 2022-06-14 | | Release date: | 2022-09-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Basis of Substrate Promiscuity and Catalysis by the Reverse Prenyltransferase N -Dimethylallyl-l-tryptophan Synthase from Fusarium fujikuroi .
Biochemistry, 61, 2022
|
|
8DB1
 
 | | Crystal structure of native DMATS1 prenyltransferase | | Descriptor: | Dimethylallyltryptophan synthase 1, TRYPTOPHAN | | Authors: | Eaton, S.A, Ronnebaum, T.A, Roose, B.W, Christianson, D.W. | | Deposit date: | 2022-06-14 | | Release date: | 2022-09-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structural Basis of Substrate Promiscuity and Catalysis by the Reverse Prenyltransferase N -Dimethylallyl-l-tryptophan Synthase from Fusarium fujikuroi .
Biochemistry, 61, 2022
|
|
2J8C
 
 | | X-ray high resolution structure of the photosynthetic reaction center from Rb. sphaeroides at pH 8 in the neutral state | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, ... | | Authors: | Koepke, J, Diehm, R, Fritzsch, G. | | Deposit date: | 2006-10-24 | | Release date: | 2007-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Ph Modulates the Quinone Position in the Photosynthetic Reaction Center from Rhodobacter Sphaeroides in the Neutral and Charge Separated States.
J.Mol.Biol., 371, 2007
|
|
1MQV
 
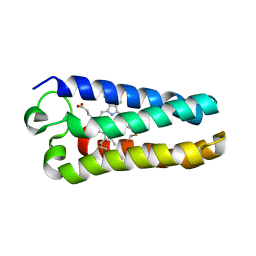 | | Crystal Structure of the Q1A/F32W/W72F mutant of Rhodopseudomonas palustris cytochrome c' (prime) expressed in E. coli | | Descriptor: | CYTOCHROME C', HEME C | | Authors: | Lee, J.C, Engman, K.C, Tezcan, F.A, Gray, H.B, Winkler, J.R. | | Deposit date: | 2002-09-17 | | Release date: | 2002-11-20 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural Features of Cytochrome c' Folding Intermediates Revealed by Fluorescence Energy-Transfer Kinetics
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
2MAD
 
 | |
7TD0
 
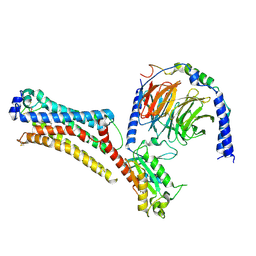 | | Lysophosphatidic acid receptor 1-Gi complex bound to LPA | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, S, Paknejad, N, Zhu, L, Kihara, Y, Ray, D, Chun, J, Liu, W, Hite, R.K, Huang, X.Y. | | Deposit date: | 2021-12-30 | | Release date: | 2022-02-09 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Differential activation mechanisms of lipid GPCRs by lysophosphatidic acid and sphingosine 1-phosphate.
Nat Commun, 13, 2022
|
|
7TD2
 
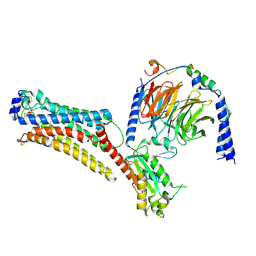 | | Lysophosphatidic acid receptor 1-Gi complex bound to LPA, state a | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, S, Paknejad, N, Zhu, L, Kihara, Y, Ray, D, Chun, J, Liu, W, Hite, R.K, Huang, X.Y. | | Deposit date: | 2021-12-30 | | Release date: | 2022-02-09 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Differential activation mechanisms of lipid GPCRs by lysophosphatidic acid and sphingosine 1-phosphate.
Nat Commun, 13, 2022
|
|
7TD3
 
 | | Sphingosine-1-phosphate receptor 1-Gi complex bound to S1P | | Descriptor: | (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liu, S, Paknejad, N, Zhu, L, Kihara, Y, Ray, D, Chun, J, Liu, W, Hite, R.K, Huang, X.Y. | | Deposit date: | 2021-12-30 | | Release date: | 2022-02-09 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Differential activation mechanisms of lipid GPCRs by lysophosphatidic acid and sphingosine 1-phosphate.
Nat Commun, 13, 2022
|
|
7TD1
 
 | | Lysophosphatidic acid receptor 1-Gi complex bound to LPA, state a | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, S, Paknejad, N, Zhu, L, Kihara, Y, Ray, D, Chun, J, Liu, W, Hite, R.K, Huang, X.Y. | | Deposit date: | 2021-12-30 | | Release date: | 2022-02-09 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Differential activation mechanisms of lipid GPCRs by lysophosphatidic acid and sphingosine 1-phosphate.
Nat Commun, 13, 2022
|
|
7TD4
 
 | | Sphingosine-1-phosphate receptor 1-Gi complex bound to Siponimod | | Descriptor: | 1-[[4-[(~{E})-~{N}-[[4-cyclohexyl-3-(trifluoromethyl)phenyl]methoxy]-~{C}-methyl-carbonimidoyl]-2-ethyl-phenyl]methyl]azetidine-3-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liu, S, Paknejad, N, Zhu, L, Kihara, Y, Ray, D, Chun, J, Liu, W, Hite, R.K, Huang, X.Y. | | Deposit date: | 2021-12-30 | | Release date: | 2022-02-09 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Differential activation mechanisms of lipid GPCRs by lysophosphatidic acid and sphingosine 1-phosphate.
Nat Commun, 13, 2022
|
|
7T6S
 
 | | Structure of the human FPR2-Gi complex with compound C43 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhuang, Y.W. | | Deposit date: | 2021-12-14 | | Release date: | 2022-03-30 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular recognition of formylpeptides and diverse agonists by the formylpeptide receptors FPR1 and FPR2.
Nat Commun, 13, 2022
|
|
7T6V
 
 | | Structure of the human FPR2-Gi complex with fMLFII | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhuang, Y.W. | | Deposit date: | 2021-12-14 | | Release date: | 2022-03-30 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular recognition of formylpeptides and diverse agonists by the formylpeptide receptors FPR1 and FPR2.
Nat Commun, 13, 2022
|
|
