5VPO
 
 | | The 70S P-site ASL SufA6 complex | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Hong, S, Sunita, S, Dunkle, J.A, Maehigashi, T, Dunham, C.M. | | Deposit date: | 2017-05-05 | | Release date: | 2018-09-26 | | Last modified: | 2018-11-07 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | Mechanism of tRNA-mediated +1 ribosomal frameshifting.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5VJX
 
 | | Crystal structure of the CLOCK Transcription Domain Exon19 in Complex with a Repressor | | Descriptor: | CLOCK-interacting pacemaker, Circadian locomoter output cycles protein kaput | | Authors: | Hou, Z, Su, L, Pei, J, Grishin, N.V, Zhang, H. | | Deposit date: | 2017-04-20 | | Release date: | 2017-12-06 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (2.695 Å) | | Cite: | Crystal Structure of the CLOCK Transactivation Domain Exon19 in Complex with a Repressor.
Structure, 25, 2017
|
|
7N32
 
 | | protofilaments of microtubule doublets bound to outer-arm dynein | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Rao, Q, Zhang, K. | | Deposit date: | 2021-05-31 | | Release date: | 2021-09-29 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structures of outer-arm dynein array on microtubule doublet reveal a motor coordination mechanism.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7NA6
 
 | | Cryo-EM structure of AAV True Type | | Descriptor: | Capsid protein VP1 | | Authors: | Bennett, A.D, McKenna, R. | | Deposit date: | 2021-06-19 | | Release date: | 2021-09-29 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Comparative structural, biophysical, and receptor binding study of true type and wild type AAV2.
J.Struct.Biol., 213, 2021
|
|
7XL8
 
 | | Human Cx36/GJD2 (N-terminal deletion mutant) gap junction channel in soybean lipids (D6 symmetry) | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction delta-2 protein | | Authors: | Lee, S.N, Cho, H.J, Jeong, H, Ryu, B, Lee, H.J, Lee, H.H, Woo, J.S. | | Deposit date: | 2022-04-21 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structures of human Cx36/GJD2 neuronal gap junction channel.
Nat Commun, 14, 2023
|
|
7MUV
 
 | | Reconstruction of the Legionella pneumophila Dot/Icm T4SS 3DVA Map 3 | | Descriptor: | DUF2807 domain-containing protein, DotC, DotD, ... | | Authors: | Sheedlo, M.J, Durie, C.L, Swanson, M, Lacy, D.B, Ohi, M.D. | | Deposit date: | 2021-05-14 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Cryo-EM reveals new species-specific proteins and symmetry elements in the Legionella pneumophila Dot/Icm T4SS.
Elife, 10, 2021
|
|
7MUY
 
 | | Reconstruction of the Legionella pneumophila Dot/Icm T4SS 3DVA Map 5 | | Descriptor: | DUF2807 domain-containing protein, DotC, DotD, ... | | Authors: | Sheedlo, M.J, Durie, C.L, Swanson, M, Lacy, D.B, Ohi, M.D. | | Deposit date: | 2021-05-14 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Cryo-EM reveals new species-specific proteins and symmetry elements in the Legionella pneumophila Dot/Icm T4SS.
Elife, 10, 2021
|
|
7MUQ
 
 | | Reconstruction of the Legionella pneumophila Dot/Icm T4SS 3DVA Map 1 | | Descriptor: | DUF2807 domain-containing protein, DotC, DotD, ... | | Authors: | Sheedlo, M.J, Durie, C.L, Swanson, M, Lacy, D.B, Ohi, M.D. | | Deposit date: | 2021-05-14 | | Release date: | 2021-10-06 | | Last modified: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Cryo-EM reveals new species-specific proteins and symmetry elements in the Legionella pneumophila Dot/Icm T4SS.
Elife, 10, 2021
|
|
7MUW
 
 | | Reconstruction of the Legionella pneumophila Dot/Icm T4SS 3DVA Map 4 | | Descriptor: | DUF2807 domain-containing protein, DotC, DotD, ... | | Authors: | Sheedlo, M.J, Durie, C.L, Swanson, M, Lacy, D.B, Ohi, M.D. | | Deposit date: | 2021-05-14 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Cryo-EM reveals new species-specific proteins and symmetry elements in the Legionella pneumophila Dot/Icm T4SS.
Elife, 10, 2021
|
|
7MUS
 
 | | Reconstruction of the Legionella pneumophila Dot/Icm T4SS 3DVA Map 2 | | Descriptor: | DUF2807 domain-containing protein, DotC, DotD, ... | | Authors: | Sheedlo, M.J, Durie, C.L, Swanson, M, Lacy, D.B, Ohi, M.D. | | Deposit date: | 2021-05-14 | | Release date: | 2021-10-06 | | Last modified: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Cryo-EM reveals new species-specific proteins and symmetry elements in the Legionella pneumophila Dot/Icm T4SS.
Elife, 10, 2021
|
|
7MUD
 
 | | Legionella pneumophila Dot/Icm T4SS OMC | | Descriptor: | DUF2807 domain-containing protein, DotC, DotD, ... | | Authors: | Sheedlo, M.J, Durie, C.L, Swanson, M, Lacy, D.B, Ohi, M.D. | | Deposit date: | 2021-05-14 | | Release date: | 2021-10-06 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM reveals new species-specific proteins and symmetry elements in the Legionella pneumophila Dot/Icm T4SS.
Elife, 10, 2021
|
|
7MUE
 
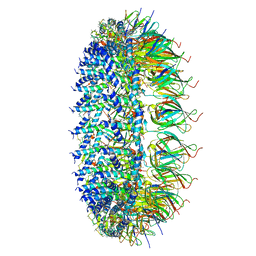 | | Legionella pneumophila Dot/Icm T4SS PR | | Descriptor: | DotF, IcmE protein, Type IV secretion protein IcmK, ... | | Authors: | Sheedlo, M.J, Durie, C.L, Swanson, M, Lacy, D.B, Ohi, M.D. | | Deposit date: | 2021-05-14 | | Release date: | 2021-10-06 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM reveals new species-specific proteins and symmetry elements in the Legionella pneumophila Dot/Icm T4SS.
Elife, 10, 2021
|
|
7MUC
 
 | | Legionella pneumophila Dot/Icm T4SS C1 Reconstruction | | Descriptor: | DUF2807 domain-containing protein, DotC, DotD, ... | | Authors: | Sheedlo, M.J, Durie, C.L, Swanson, M, Lacy, D.B, Ohi, M.D. | | Deposit date: | 2021-05-14 | | Release date: | 2021-10-06 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM reveals new species-specific proteins and symmetry elements in the Legionella pneumophila Dot/Icm T4SS.
Elife, 10, 2021
|
|
5VYC
 
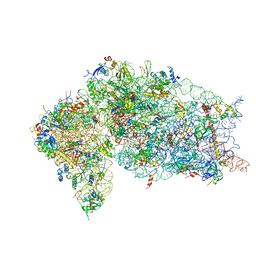 | | Crystal structure of the human 40S ribosomal subunit in complex with DENR-MCT-1. | | Descriptor: | 40S ribosomal protein S10, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | Authors: | Lomakin, I.B, Stolboushkina, E.A, Vaidya, A.T, Garber, M.B, Dmitriev, S.E, Steitz, T.A. | | Deposit date: | 2017-05-24 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (6 Å) | | Cite: | Crystal Structure of the Human Ribosome in Complex with DENR-MCT-1.
Cell Rep, 20, 2017
|
|
5VKU
 
 | | An atomic structure of the human cytomegalovirus (HCMV) capsid with its securing layer of pp150 tegument protein | | Descriptor: | Major capsid protein, Small capsomere-interacting protein, Tegument protein pp150, ... | | Authors: | Yu, X, Jih, J, Jiang, J, Zhou, H. | | Deposit date: | 2017-04-24 | | Release date: | 2017-06-28 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Atomic structure of the human cytomegalovirus capsid with its securing tegument layer of pp150.
Science, 356, 2017
|
|
7XO4
 
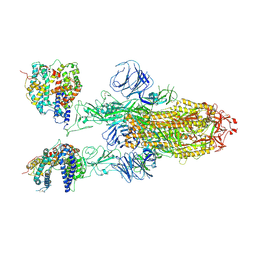 | | SARS-CoV-2 Omicron BA.1 Variant Spike Trimer with two mouse ACE2 Bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike glycoprotein, ... | | Authors: | Xu, Y, Wu, C, Liu, H, Yin, W, Xu, H.E. | | Deposit date: | 2022-04-30 | | Release date: | 2022-06-15 | | Last modified: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Structural and biochemical mechanism for increased infectivity and immune evasion of Omicron BA.2 variant compared to BA.1 and their possible mouse origins.
Cell Res., 32, 2022
|
|
7NAS
 
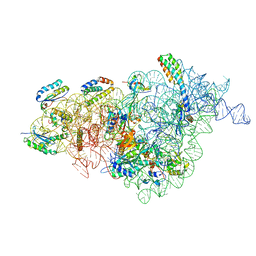 | | Bacterial 30S ribosomal subunit assembly complex state A (multibody refinement for body domain of 30S ribosome) | | Descriptor: | 16S rRNA, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S. | | Deposit date: | 2021-01-25 | | Release date: | 2021-07-07 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
7N1Q
 
 | | Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Cai, Y.F, Xiao, T.S, Rawson, S, Peng, H.Q, Sterling, S.M, Walsh Jr, R.M, Volloch, S.R, Chen, B. | | Deposit date: | 2021-05-28 | | Release date: | 2021-07-07 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants.
Science, 373, 2021
|
|
7MWM
 
 | | Crystal structure of MBD2 with DNA | | Descriptor: | DNA (5'-D(*GP*CP*CP*AP*A)-R(P*(5MC))-D(P*GP*TP*TP*GP*GP*C)-3'), Methyl-CpG-binding domain protein 2, UNKNOWN ATOM OR ION | | Authors: | Liu, K, Dong, A, Edwards, A.M, Arrowsmith, C.H, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-05-17 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Crystal structure of MBD2 with DNA
To Be Published
|
|
7XOB
 
 | | SARS-CoV-2 Omicron BA.2 Variant Spike Trimer with two mouse ACE2 Bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike glycoprotein, ... | | Authors: | Xu, Y, Wu, C, Liu, H, Yin, W, Xu, H.E. | | Deposit date: | 2022-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural and biochemical mechanism for increased infectivity and immune evasion of Omicron BA.2 variant compared to BA.1 and their possible mouse origins.
Cell Res., 32, 2022
|
|
7XO5
 
 | | SARS-CoV-2 Omicron BA.1 Variant Spike Trimer with one mouse ACE2 Bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike glycoprotein, ... | | Authors: | Xu, Y, Wu, C, Liu, H, Yin, W, Xu, H.E. | | Deposit date: | 2022-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structural and biochemical mechanism for increased infectivity and immune evasion of Omicron BA.2 variant compared to BA.1 and their possible mouse origins.
Cell Res., 32, 2022
|
|
7XO7
 
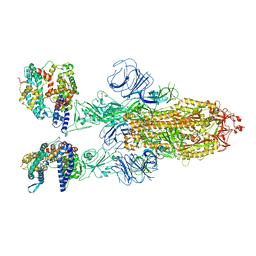 | | SARS-CoV-2 Omicron BA.2 Variant Spike Trimer with two human ACE2 Bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, CHLORIDE ION, ... | | Authors: | Xu, Y, Wu, C, Liu, H, Yin, W, Xu, H.E. | | Deposit date: | 2022-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structural and biochemical mechanism for increased infectivity and immune evasion of Omicron BA.2 variant compared to BA.1 and their possible mouse origins.
Cell Res., 32, 2022
|
|
7N1U
 
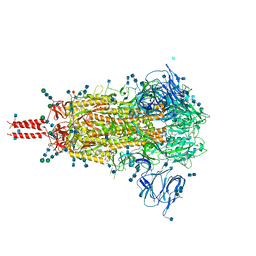 | | Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Cai, Y.F, Xiao, T.S, Rawson, S, Peng, H.Q, Sterling, S.M, Walsh Jr, R.M, Volloch, S.R, Chen, B. | | Deposit date: | 2021-05-28 | | Release date: | 2021-07-07 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants.
Science, 373, 2021
|
|
7XO8
 
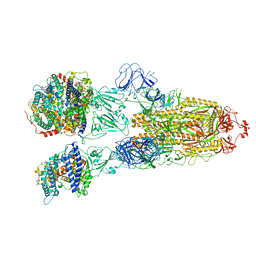 | | SARS-CoV-2 Omicron BA.2 Variant Spike Trimer with three human ACE2 Bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, CHLORIDE ION, ... | | Authors: | Xu, Y, Wu, C, Liu, H, Yin, W, Xu, H.E. | | Deposit date: | 2022-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structural and biochemical mechanism for increased infectivity and immune evasion of Omicron BA.2 variant compared to BA.1 and their possible mouse origins.
Cell Res., 32, 2022
|
|
7N1X
 
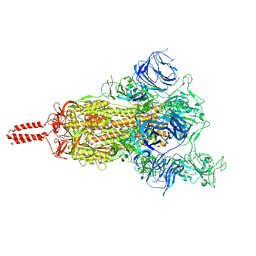 | | Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Cai, Y.F, Xiao, T.S, Rawson, S, Peng, H.Q, Sterling, S.M, Walsh Jr, R.M, Volloch, S.R, Chen, B. | | Deposit date: | 2021-05-28 | | Release date: | 2021-07-07 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants.
Science, 373, 2021
|
|
