4RSG
 
 | | Neutron crystal structure of Ras bound to the GTP analogue GppNHp | | Descriptor: | GTPase HRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Knihtila, R.R, Holzapfel, G, Weiss, K.L, Meilleur, F, Mattos, C. | | Deposit date: | 2014-11-07 | | Release date: | 2015-11-04 | | Last modified: | 2024-02-28 | | Method: | NEUTRON DIFFRACTION (1.907 Å) | | Cite: | Neutron Crystal Structure of RAS GTPase Puts in Question the Protonation State of the GTP gamma-Phosphate.
J.Biol.Chem., 290, 2015
|
|
7ACQ
 
 | | CRYSTAL STRUCTURE OF INACTIVE KRAS G12D (GDP) IN COMPLEX WITH THE SOAKED DIMERIC INHIBITOR BI-5747 | | Descriptor: | (3~{S})-5-oxidanyl-3-[2-[[6-[[3-[(1~{S})-6-oxidanyl-3-oxidanylidene-1,2-dihydroisoindol-1-yl]-1~{H}-indol-2-yl]methylamino]hexylamino]methyl]-1~{H}-indol-3-yl]-2,3-dihydroisoindol-1-one, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Kessler, D. | | Deposit date: | 2020-09-11 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | CRYSTAL STRUCTURE OF INACTIVE KRAS G12D (GDP) IN COMPLEX WITH THE SOAKED DIMERIC INHIBITOR BI00925747
To Be Published
|
|
6GZM
 
 | | Crystal Structure of Human CKIdelta with A86 | | Descriptor: | CITRIC ACID, Casein kinase I isoform delta, GLYCEROL, ... | | Authors: | Ben-neriah, Y, Venkatachalam, A, Minzel, W, Fink, A, Snir-Alkalay, I, Vacca, J. | | Deposit date: | 2018-07-04 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Small Molecules Co-targeting CKI alpha and the Transcriptional Kinases CDK7/9 Control AML in Preclinical Models.
Cell, 175, 2018
|
|
4RTZ
 
 | |
6GHV
 
 | | Structure of a DC-SIGN CRD in complex with high affinity glycomimetic. | | Descriptor: | CALCIUM ION, CD209 antigen, CHLORIDE ION, ... | | Authors: | Thepaut, M, Achilli, S, Medve, L, Bernardi, A, Fieschi, F. | | Deposit date: | 2018-05-09 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Enhancing Potency and Selectivity of a DC-SIGN Glycomimetic Ligand by Fragment-Based Design: Structural Basis.
Chemistry, 25, 2019
|
|
7BS8
 
 | | Bovine Pancreatic Trypsin with Benzamidine (Cryo) | | Descriptor: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Maeki, M, Ito, S, Takeda, R, Funakubo, T, Ueno, G, Ishida, A, Tani, H, Yamamoto, M, Tokeshi, M. | | Deposit date: | 2020-03-30 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Room-temperature crystallography using a microfluidic protein crystal array device and its application to protein-ligand complex structure analysis.
Chem Sci, 11, 2020
|
|
6GII
 
 | | The crystal structure of Tepidiphilus thermophilus P450 heme domain | | Descriptor: | Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Levy, C.W. | | Deposit date: | 2018-05-11 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of P450-TT heme-domain provides the first structural insights into the versatile class VII P450s.
Biochem. Biophys. Res. Commun., 501, 2018
|
|
4RW1
 
 | | Hen egg-white lysozyme structure from a spent-beam experiment at LCLS: original beam | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Boutet, S, Foucar, L, Botha, S, Doak, R.B, Koglin, J.E, Messerschmidt, M, Nass, K, Schlichting, I, Shoeman, R, Williams, G.J. | | Deposit date: | 2014-12-01 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterization and use of the spent beam for serial operation of LCLS.
J.SYNCHROTRON RADIAT., 22, 2015
|
|
4ROB
 
 | | 2.8A resolution structure of SRPN2 (K198C) from Anopheles gambiae | | Descriptor: | Serpin 2 | | Authors: | Lovell, S, Battaile, K.P, Zhang, X, Meekins, D.A, An, C, Michel, K. | | Deposit date: | 2014-10-28 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | 2.8A resolution structure of SRPN2 (K198C) from Anopheles gambiae
To be Published
|
|
5FXL
 
 | |
5F94
 
 | | Crystal structure of GSK3b in complex with Compound 15: 2-[(cyclopropylcarbonyl)amino]-N-(4-methoxypyridin-3-yl)pyridine-4-carboxamide | | Descriptor: | 2-[(cyclopropylcarbonyl)amino]-N-(4-methoxypyridin-3-yl)pyridine-4-carboxamide, Glycogen synthase kinase-3 beta | | Authors: | Lewis, H.A, Kish, K, Luo, G, Dubowchick, G.M. | | Deposit date: | 2015-12-09 | | Release date: | 2016-02-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Discovery of Isonicotinamides as Highly Selective, Brain Penetrable, and Orally Active Glycogen Synthase Kinase-3 Inhibitors.
J.Med.Chem., 59, 2016
|
|
7C0P
 
 | |
4RTW
 
 | |
8Q3L
 
 | |
7B8H
 
 | | Monoclinic structure of human protein kinase CK2 catalytic subunit in complex with a heparin oligo saccharide | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, Casein kinase II subunit alpha, GLYCEROL, ... | | Authors: | Niefind, K, Schnitzler, A. | | Deposit date: | 2020-12-12 | | Release date: | 2021-02-03 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Structural basis for the design of bisubstrate inhibitors of protein kinase CK2 provided by complex structures with the substrate-competitive inhibitor heparin.
Eur.J.Med.Chem., 214, 2021
|
|
7BPS
 
 | | Crystal structure of mouse TEX101 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, Testis-expressed protein 101, ... | | Authors: | Masutani, M, Sakurai, S, Shimizu, T, Ohto, U. | | Deposit date: | 2020-03-23 | | Release date: | 2020-07-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of TEX101, a glycoprotein essential for male fertility, reveals the presence of tandemly arranged Ly6/uPAR domains.
Febs Lett., 594, 2020
|
|
6HBK
 
 | | Echovirus 18 Open particle without one pentamer | | Descriptor: | Echovirus 18 capsid protein 1, Echovirus 18 capsid protein 2, Echovirus 18 capsid protein 3 | | Authors: | Buchta, D, Fuzik, T, Hrebik, D, Levdansky, Y, Moravcova, J, Plevka, P. | | Deposit date: | 2018-08-10 | | Release date: | 2019-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Enterovirus particles expel capsid pentamers to enable genome release.
Nat Commun, 10, 2019
|
|
7BS2
 
 | | Bovine Pancreatic Trypsin with serotonin (Room Temperature) | | Descriptor: | CALCIUM ION, Cationic trypsin, SEROTONIN, ... | | Authors: | Maeki, M, Ito, S, Takeda, R, Funakubo, T, Ueno, G, Ishida, A, Tani, H, Yamamoto, M, Tokeshi, M. | | Deposit date: | 2020-03-30 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Room-temperature crystallography using a microfluidic protein crystal array device and its application to protein-ligand complex structure analysis.
Chem Sci, 11, 2020
|
|
8B49
 
 | | STRUCTURE OF PORCINE PANCREATIC ELASTASE BOUND TO A FRAGMENT (m-toluoylcarbonyl group) OF A 5-AZAINDOLE INHIBITOR | | Descriptor: | 1-(3-methylphenyl)carbonylpyrrolo[3,2-c]pyridine-3-carbonitrile, CALCIUM ION, Chymotrypsin-like elastase family member 1, ... | | Authors: | Ferraroni, M, Giovannoni, P, Gerace, A. | | Deposit date: | 2022-09-20 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | X-ray structural study of human neutrophil elastase inhibition with a series of azaindoles, azaindazoles and isoxazolones
J.Mol.Struct., 1274, 2023
|
|
6TVY
 
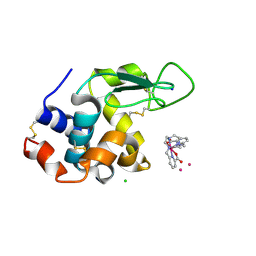 | | Structure of hen egg white lysozyme crystallized in the presence of Tb-Xo4 crystallophore in the XtalController device | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION, ... | | Authors: | de Wijn, R, Rollet, K, Coudray, L, McEwen, A.G, Lorber, B, Sauter, C. | | Deposit date: | 2020-01-10 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Monitoring the Production of High Diffraction-Quality Crystals of Two Enzymes in Real Time Using In Situ Dynamic Light Scattering
Crystals, 2020
|
|
4S33
 
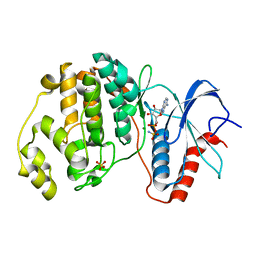 | | ERK2 R65S mutant complexed with AMP-PNP | | Descriptor: | MAGNESIUM ION, Mitogen-activated protein kinase 1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Livnah, O, Karamansha, Y, Tzarum, N. | | Deposit date: | 2015-01-26 | | Release date: | 2016-01-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Multiple mechanisms render Erk proteins MEK-independent
To be Published
|
|
5FFS
 
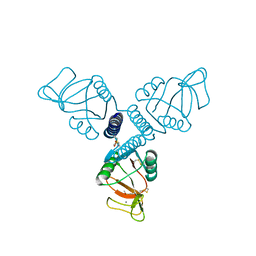 | | Crystal Structure of Surfactant Protein-A Y164A Mutant | | Descriptor: | CALCIUM ION, Pulmonary surfactant-associated protein A | | Authors: | Goh, B.C, Wu, H, Rynkiewicz, M.J, Schulten, K, Seaton, B.A, McCormack, F.X. | | Deposit date: | 2015-12-18 | | Release date: | 2016-07-06 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Elucidation of Lipid Binding Sites on Lung Surfactant Protein A Using X-ray Crystallography, Mutagenesis, and Molecular Dynamics Simulations.
Biochemistry, 55, 2016
|
|
7BF8
 
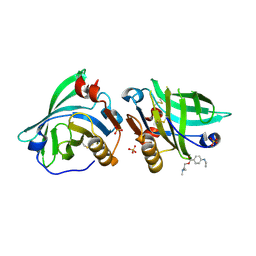 | |
8BO4
 
 | | COAGULATION FACTOR XI PROTEASE DOMAIN IN COMPLEX WITH ACTIVE SITE INHIBITOR 1 | | Descriptor: | 4-(aminomethyl)-~{N}-[(2~{S})-1-oxidanylidene-3-phenyl-1-[[4-(1~{H}-1,2,3,4-tetrazol-5-yl)phenyl]amino]propan-2-yl]cyclohexane-1-carboxamide, Coagulation factor XIa light chain, GLYCEROL | | Authors: | Schaefer, M, Roehrig, S, Ackerstaff, J, Nunez, E.J, Gericke, K.M, Meier, K, Tersteegen, A, Stampfuss, J, Ellerbrock, P, Meibom, M, Lang, D, Heitmeier, S, Hillisch, A. | | Deposit date: | 2022-11-14 | | Release date: | 2023-09-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Design and Preclinical Characterization Program toward Asundexian (BAY 2433334), an Oral Factor XIa Inhibitor for the Prevention and Treatment of Thromboembolic Disorders.
J.Med.Chem., 66, 2023
|
|
8BO3
 
 | | COAGULATION FACTOR XI PROTEASE DOMAIN IN COMPLEX WITH ACTIVE SITE INHIBITOR Asundexian | | Descriptor: | 4-[[(2~{S})-2-[4-[5-chloranyl-2-[4-(trifluoromethyl)-1,2,3-triazol-1-yl]phenyl]-5-methoxy-2-oxidanylidene-pyridin-1-yl]butanoyl]amino]-2-fluoranyl-benzamide, CITRIC ACID, Coagulation factor XIa light chain | | Authors: | Schaefer, M, Roehrig, S, Ackerstaff, J, Nunez, E.J, Gericke, K.M, Meier, K, Tersteegen, A, Stampfuss, J, Ellerbrock, P, Meibom, D, Lang, D, Heitmeier, S, Hillisch, A. | | Deposit date: | 2022-11-14 | | Release date: | 2023-09-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.841 Å) | | Cite: | Design and Preclinical Characterization Program toward Asundexian (BAY 2433334), an Oral Factor XIa Inhibitor for the Prevention and Treatment of Thromboembolic Disorders.
J.Med.Chem., 66, 2023
|
|
