6JE8
 
 | | crystal structure of a beta-N-acetylhexosaminidase | | Descriptor: | Beta-N-acetylhexosaminidase, FORMIC ACID, GLYCEROL, ... | | Authors: | Chen, X, Wang, J.C, Liu, M.J, Yang, W.Y, Wang, Y.Z, Tang, R.P, Zhang, M. | | Deposit date: | 2019-02-04 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Crystallographic evidence for substrate-assisted catalysis of beta-N-acetylhexosaminidas from Akkermansia muciniphila.
Biochem. Biophys. Res. Commun., 511, 2019
|
|
2EPM
 
 | | N-acetyl-B-D-glucoasminidase (GCNA) from Stretococcus gordonii | | Descriptor: | GLYCEROL, MERCURY (II) ION, N-acetyl-beta-D-glucosaminidase, ... | | Authors: | Langley, D.B, Harty, D.W.S, Guss, J.M, Collyer, C.A. | | Deposit date: | 2007-03-30 | | Release date: | 2008-03-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure of N-acetyl-beta-D-glucosaminidase (GcnA) from the Endocarditis Pathogen Streptococcus gordonii and its Complex with the Mechanism-based Inhibitor NAG-thiazoline
J.Mol.Biol., 377, 2008
|
|
2R5V
 
 | | Hydroxymandelate Synthase Crystal Structure | | Descriptor: | (2S)-hydroxy(4-hydroxyphenyl)ethanoic acid, COBALT (II) ION, PCZA361.1, ... | | Authors: | Brownlee, J.M, He, P, Moran, G.R, Harrison, D.H.T. | | Deposit date: | 2007-09-04 | | Release date: | 2008-03-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Two roads diverged: the structure of hydroxymandelate synthase from Amycolatopsis orientalis in complex with 4-hydroxymandelate.
Biochemistry, 47, 2008
|
|
4MAZ
 
 | | The Structure of MalL mutant enzyme V200S from Bacillus subtilus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Hobbs, J.K, Jiao, W, Easter, A.D, Parker, E.J, Schipper, L.A, Arcus, V.L. | | Deposit date: | 2013-08-18 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Change in heat capacity for enzyme catalysis determines temperature dependence of enzyme catalyzed rates.
Acs Chem.Biol., 8, 2013
|
|
6JGC
 
 | | Crystal structure of barley exohydrolaseI W286Y mutant in complex with glucose. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-02-13 | | Release date: | 2020-08-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
4QLL
 
 | | Crystal structure of rice BGlu1 E176Q/Y341A/Q187A mutant complexed with cellotetraose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase 7, SULFATE ION, ... | | Authors: | Pengthaisong, S, Ketudat Cairns, J.R. | | Deposit date: | 2014-06-12 | | Release date: | 2015-06-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Effects of active site cleft residues on oligosaccharide binding, hydrolysis, and glycosynthase activities of rice BGlu1 and its mutants
Protein Sci., 23, 2014
|
|
6JG7
 
 | | Crystal structure of barley exohydrolaseI W286F in complex with methyl 2-thio-beta-sophoroside | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1, GLYCEROL, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-02-13 | | Release date: | 2020-08-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
2EQ9
 
 | |
3HS5
 
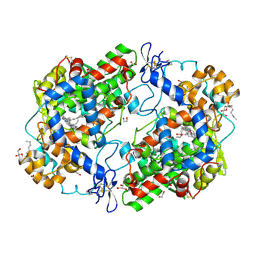 | | X-ray crystal structure of arachidonic acid bound to the cyclooxygenase channel of cyclooxygenase-2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Vecchio, A.J, Simmons, D.M, Malkowski, M.G. | | Deposit date: | 2009-06-10 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of fatty acid substrate binding to cyclooxygenase-2.
J.Biol.Chem., 285, 2010
|
|
3DGH
 
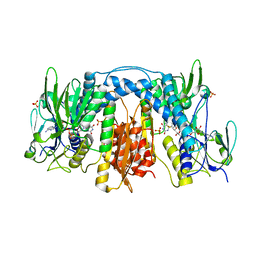 | | Crystal Structure of Drosophila Thioredoxin Reductase, C-terminal 8-residue truncation | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, Thioredoxin reductase 1, ... | | Authors: | Eckenroth, B.E, Hondal, R.J, Everse, S.J. | | Deposit date: | 2008-06-13 | | Release date: | 2009-06-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.745 Å) | | Cite: | Crystal Structure of Drosophila Thioredoxin Reductase, C-terminal 8-residue truncation
To be Published
|
|
6JGS
 
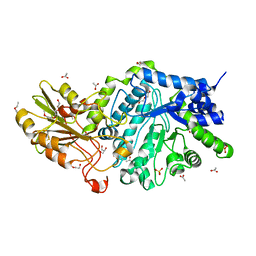 | | Crystal structure of barley exohydrolaseI W434Y mutant in complex with 4I,4III,4V-S-trithiocellohexaose. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-02-14 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
3DHU
 
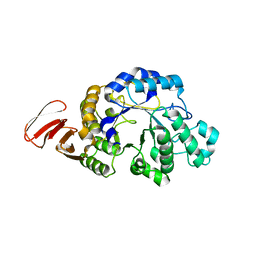 | | Crystal structure of an alpha-amylase from Lactobacillus plantarum | | Descriptor: | Alpha-amylase | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Iizuka, M, Ozyurt, S, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-06-18 | | Release date: | 2008-08-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of an alpha-amylase from Lactobacillus plantarum
To be Published
|
|
6JHG
 
 | | Crystal structure of apo Pullulanase from Paenibacillus barengoltzii in space group P212121 | | Descriptor: | CALCIUM ION, CHLORIDE ION, Pulullanase | | Authors: | Wu, S.W, Yang, S.Q, Qin, Z, You, X, Huang, P, Jiang, Z.Q. | | Deposit date: | 2019-02-18 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.891 Å) | | Cite: | Crystal structure of apo Pullulanase from Paenibacillus barengoltzii in space group P212121
To Be Published
|
|
1YQE
 
 | | Crystal Structure of Conserved Protein of Unknown Function AF0625 | | Descriptor: | Hypothetical UPF0204 protein AF0625, PYROPHOSPHATE 2- | | Authors: | Liu, Y, Skarina, T, Dong, A, Kudritskam, M, Savchenko, A, Pai, E.F, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-02-01 | | Release date: | 2005-03-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal Structure of Conserved Hypothetical Protein AF0625
To be Published
|
|
3HQ4
 
 | | Crystal Structure of C151S mutant of Glyceraldehyde-3-phosphate dehydrogenase 1 (GAPDH1) complexed with NAD from Staphylococcus aureus MRSA252 at 2.2 angstrom resolution | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase 1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Mukherjee, S, Dutta, D, Saha, B, Das, A.K. | | Deposit date: | 2009-06-05 | | Release date: | 2010-06-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase 1 from methicillin-resistant Staphylococcus aureus MRSA252 provides novel insights into substrate binding and catalytic mechanism.
J.Mol.Biol., 401, 2010
|
|
2R7J
 
 | |
4I1M
 
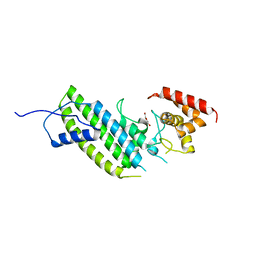 | | Crystal structure of the Legionella pneumophila GAP domain of LepB | | Descriptor: | DI(HYDROXYETHYL)ETHER, LepB | | Authors: | Streller, A, Gazdag, E.M, Vetter, I.R, Goody, R.S, Itzen, A. | | Deposit date: | 2012-11-21 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | Mechanism of Rab1b deactivation by the Legionella pneumophila GAP LepB.
Embo Rep., 14, 2013
|
|
2RB2
 
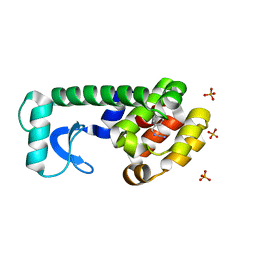 | |
6ASL
 
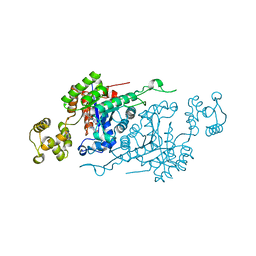 | |
1YNY
 
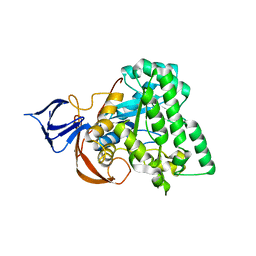 | | Molecular Structure of D-Hydantoinase from a Bacillus sp. AR9: Evidence for mercury inhibition | | Descriptor: | D-Hydantoinase, MANGANESE (II) ION | | Authors: | Radha Kishan, K.V, Vohra, R.M, Ganeshan, K, Agrawal, V, Sharma, V.M, Sharma, R. | | Deposit date: | 2005-01-26 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular structure of D-hydantoinase from Bacillus sp. AR9: evidence for mercury inhibition.
J.Mol.Biol., 347, 2005
|
|
4B56
 
 | | Structure of ectonucleotide pyrophosphatase-phosphodiesterase-1 (NPP1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Jansen, S, Perrakis, A, Ulens, C, Winkler, C, Andries, M, Joosten, R.P, Van Acker, M, Luyten, F.P, Moolenaar, W.H, Bollen, M. | | Deposit date: | 2012-08-02 | | Release date: | 2012-09-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of Npp1, an Ectonucleotide Pyrophosphatase/Phosphodiesterase Involved in Tissue Calcification.
Structure, 20, 2012
|
|
4I9B
 
 | | Structure of aminoaldehyde dehydrogenase 1 from Solanum lycopersium (SlAMADH1) with a thiohemiacetal intermediate | | Descriptor: | (2-hydroxyethoxy)acetaldehyde, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Morera, S, Vigouroux, A, Kopecny, D. | | Deposit date: | 2012-12-05 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Plant ALDH10 family: identifying critical residues for substrate specificity and trapping a thiohemiacetal intermediate.
J.Biol.Chem., 288, 2013
|
|
2EJD
 
 | | Crystal analysis of delta1-pyrroline-5-carboxylate dehydrogenase from Thermus thermophilus with bound L-alanine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-pyrroline-5-carboxylate dehydrogenase, ACETATE ION, ... | | Authors: | Inagaki, E, Sakamoto, K, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-16 | | Release date: | 2007-09-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure analysis of delta1-pyrroline-5-carboxylate dehydrogenase in ternary complex with inhibitor and NAD
To be Published
|
|
6JIT
 
 | | Complex structure of an imine reductase at 2.05 Angstrom resolution | | Descriptor: | 1-(2-phenylethyl)-3,4-dihydroisoquinoline, 6-phosphogluconate dehydrogenase NAD-binding protein, CHLORIDE ION, ... | | Authors: | Li, H, Wu, L, Zheng, G.W, Zhou, J.H. | | Deposit date: | 2019-02-23 | | Release date: | 2020-02-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | Complex structure of an imine reductase at 2.05 Angstrom resolution
To Be Published
|
|
4ATL
 
 | | Crystal structure of Raucaffricine glucosidase in complex with Glucose | | Descriptor: | RAUCAFFRICINE-O-BETA-D-GLUCOSIDASE, beta-D-glucopyranose | | Authors: | Xia, L, Rajendran, C, Ruppert, M, Panjikar, S, Wang, M, Stoeckigt, J. | | Deposit date: | 2012-05-08 | | Release date: | 2013-01-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | High Speed X-Ray Analysis of Plant Enzymes at Room Temperature
Phytochemistry, 91, 2013
|
|
