6TZ1
 
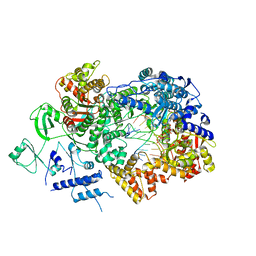 | | In situ structure of BmCPV RNA-dependent RNA polymerase at early-elongation state | | Descriptor: | Non-template RNA (5'-D(*(GTA))-R(P*GP*UP*A)-3'), RNA-dependent RNA Polymerase, Template RNA (5'-R(P*AP*GP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), ... | | Authors: | Cui, Y.X, Zhang, Y.N, Sun, J.C, Zhou, Z.H. | | Deposit date: | 2019-08-09 | | Release date: | 2019-11-20 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Conservative transcription in three steps visualized in a double-stranded RNA virus.
Nat.Struct.Mol.Biol., 26, 2019
|
|
8E27
 
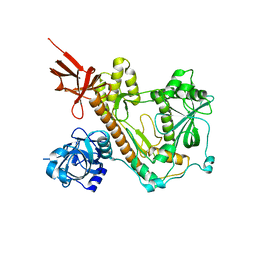 | | RNA-free Human Dis3L2 | | Descriptor: | DIS3-like exonuclease 2 | | Authors: | Meze, K, Thomas, D.R, Joshua-Tor, L. | | Deposit date: | 2022-08-14 | | Release date: | 2023-03-01 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | A shape-shifting nuclease unravels structured RNA.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6SVS
 
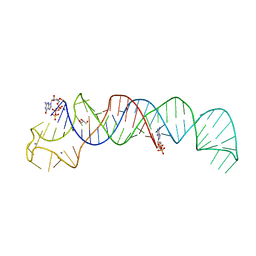 | | Crystal Structure of U:A-U-rich RNA triple helix with 11 consecutive base triples | | Descriptor: | ADENOSINE-5'-PHOSPHATE-2',3'-CYCLIC PHOSPHATE, CALCIUM ION, GLYCEROL, ... | | Authors: | Ruszkowska, A, Ruszkowski, M, Hulewicz, J.P, Dauter, Z, Brown, J.A. | | Deposit date: | 2019-09-18 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular structure of a U•A-U-rich RNA triple helix with 11 consecutive base triples.
Nucleic Acids Res., 48, 2020
|
|
7XSR
 
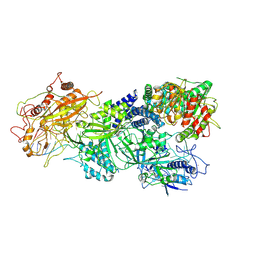 | | Structure of Craspase-target RNA | | Descriptor: | CHAT domain protein, RAMP superfamily protein, RNA (34-MER), ... | | Authors: | Feng, Y, Zhang, L. | | Deposit date: | 2022-05-15 | | Release date: | 2022-11-09 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Target RNA activates the protease activity of Craspase to confer antiviral defense.
Mol.Cell, 82, 2022
|
|
1FJE
 
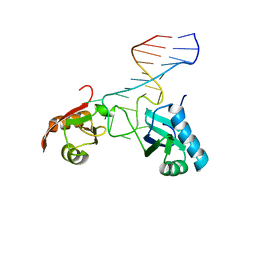 | | SOLUTION STRUCTURE OF NUCLEOLIN RBD12 IN COMPLEX WITH SNRE RNA | | Descriptor: | NUCLEOLIN RBD12, SNRE RNA | | Authors: | Allain, F.H.T, Bouvet, P, Dieckmann, T, Feigon, J. | | Deposit date: | 2000-08-08 | | Release date: | 2001-01-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Molecular basis of sequence-specific recognition of pre-ribosomal RNA by nucleolin.
EMBO J., 19, 2000
|
|
5VT0
 
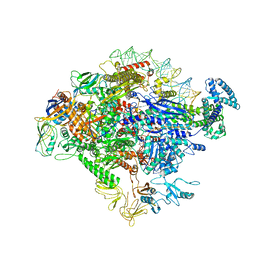 | |
2BX9
 
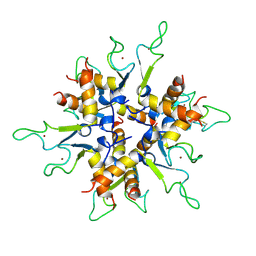 | | Crystal structure of B.subtilis Anti-TRAP protein, an antagonist of TRAP-RNA interactions | | Descriptor: | TRYPTOPHAN RNA-BINDING ATTENUATOR PROTEIN-INHIBITORY PROTEIN, ZINC ION | | Authors: | Shevtsov, M.B, Chen, Y, Gollnick, P, Antson, A.A. | | Deposit date: | 2005-07-26 | | Release date: | 2005-11-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Bacillus Subtilis Anti-Trap Protein, an Antagonist of Trap/RNA Interaction.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
4CQN
 
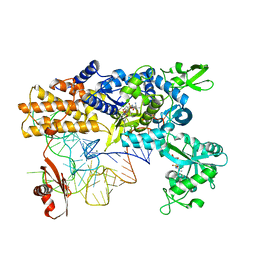 | | Crystal structure of the E.coli LeuRS-tRNA complex with the non- cognate isoleucyl adenylate analogue | | Descriptor: | ESCHERICHIA COLI TRNA-LEU UAA ISOACCEPTOR, LEUCINE--TRNA LIGASE, MAGNESIUM ION, ... | | Authors: | Palencia, A, Cusack, S, Cvetesic, N, Haslaz, I, Gruic-Sovulj, I. | | Deposit date: | 2014-02-20 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Physiological Target for Leurs Translational Quality Control is Norvaline
Embo J., 33, 2014
|
|
7Q0J
 
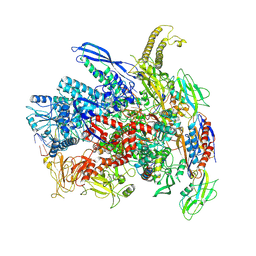 | | RNA polymerase elongation complex in more-swiveled conformation | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zhu, C, Guo, X, Weixlbaumer, A. | | Deposit date: | 2021-10-15 | | Release date: | 2022-03-23 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Transcription factors modulate RNA polymerase conformational equilibrium.
Nat Commun, 13, 2022
|
|
7Q0K
 
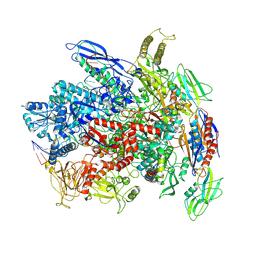 | | RNA polymerase elongation complex in less-swiveled conformation | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zhu, C, Guo, X, Weixlbaumer, A. | | Deposit date: | 2021-10-15 | | Release date: | 2022-03-23 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Transcription factors modulate RNA polymerase conformational equilibrium.
Nat Commun, 13, 2022
|
|
8S9U
 
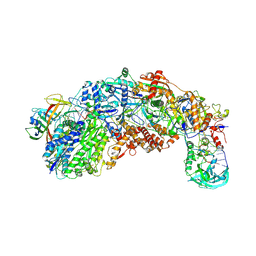 | |
8S9V
 
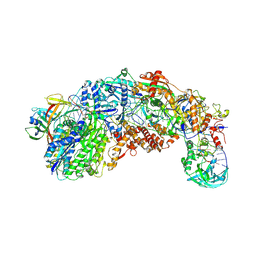 | |
8S9X
 
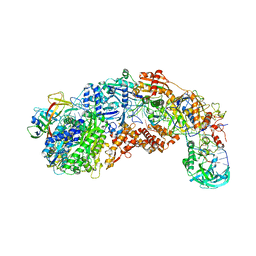 | |
5VJB
 
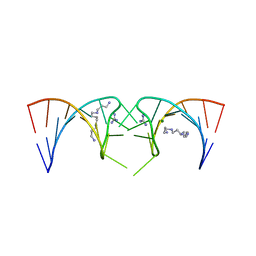 | |
2XLJ
 
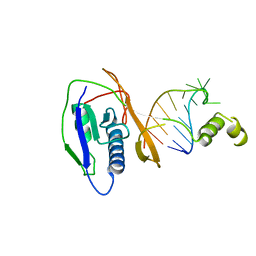 | | Crystal structure of the Csy4-crRNA complex, hexagonal form | | Descriptor: | 5'-R(*CP*UP*GP*CP*CP*GP*UP*AP*UP*AP*GP*GP*CP*A*DG*C)-3', CSY4 ENDORIBONUCLEASE | | Authors: | Haurwitz, R.E, Jinek, M, Wiedenheft, B, Zhou, K, Doudna, J.A. | | Deposit date: | 2010-07-20 | | Release date: | 2010-09-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Sequence- and Structure-Specific RNA Processing by a Crispr Endonuclease.
Science, 329, 2010
|
|
7XSP
 
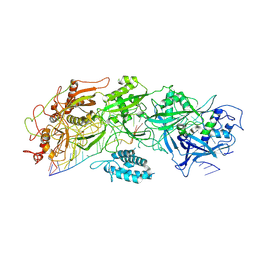 | | Structure of gRAMP-target RNA | | Descriptor: | RAMP superfamily protein, RNA (35-MER), RNA (5'-R(P*GP*GP*GP*GP*CP*AP*GP*AP*AP*AP*AP*UP*UP*GP*G)-3'), ... | | Authors: | Feng, Y, Zhang, L.X. | | Deposit date: | 2022-05-15 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Target RNA activates the protease activity of Craspase to confer antiviral defense.
Mol.Cell, 82, 2022
|
|
3G1Z
 
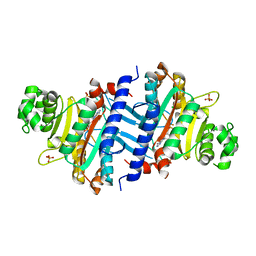 | | Structure of IDP01693/yjeA, a potential t-RNA synthetase from Salmonella typhimurium | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, PHOSPHATE ION, ... | | Authors: | Singer, A.U, Evdokimova, E, Kudritska, M, Cuff, M.E, Edwards, A.M, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-01-30 | | Release date: | 2009-03-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | PoxA, yjeK, and elongation factor P coordinately modulate virulence and drug resistance in Salmonella enterica.
Mol.Cell, 39, 2010
|
|
2XLI
 
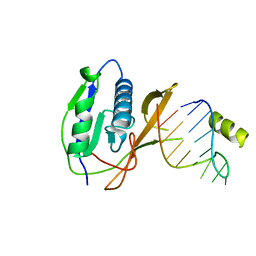 | | Crystal structure of the Csy4-crRNA complex, monoclinic form | | Descriptor: | 5'-R(*CP*UP*GP*CP*CP*GP*UP*AP*UP*AP*GP*GP*CP*A*DG*C)-3', CSY4 ENDORIBONUCLEASE | | Authors: | Haurwitz, R.E, Jinek, M, Wiedenheft, B, Zhou, K, Doudna, J.A. | | Deposit date: | 2010-07-20 | | Release date: | 2010-09-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Sequence- and Structure-Specific RNA Processing by a Crispr Endonuclease.
Science, 329, 2010
|
|
2XLK
 
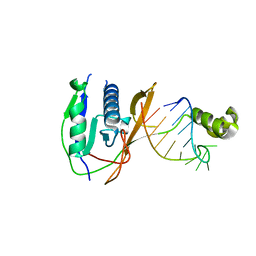 | | Crystal structure of the Csy4-crRNA complex, orthorhombic form | | Descriptor: | 5'-R(*CP*UP*GP*CP*CP*GP*UP*AP*UP*AP*GP*GP*CP*A*DG*C)-3', CSY4 ENDORIBONUCLEASE | | Authors: | Haurwitz, R.E, Jinek, M, Wiedenheft, B, Zhou, K, Doudna, J.A. | | Deposit date: | 2010-07-21 | | Release date: | 2010-09-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.805 Å) | | Cite: | Sequence- and Structure-Specific RNA Processing by a Crispr Endonuclease.
Science, 329, 2010
|
|
3N2Y
 
 | |
3K0J
 
 | | Crystal structure of the E. coli ThiM riboswitch in complex with thiamine pyrophosphate and the U1A crystallization module | | Descriptor: | MAGNESIUM ION, RNA (87-MER), THIAMINE DIPHOSPHATE, ... | | Authors: | Kulshina, N, Edwards, T.E, Ferre-D'Amare, A.R. | | Deposit date: | 2009-09-24 | | Release date: | 2009-12-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Thermodynamic analysis of ligand binding and ligand binding-induced tertiary structure formation by the thiamine pyrophosphate riboswitch.
Rna, 16, 2010
|
|
3PEW
 
 | | S. cerevisiae Dbp5 L327V bound to RNA and ADP BeF3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase DBP5, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Montpetit, B, Thomsen, N.D, Helmke, K.J, Seeliger, M.A, Berger, J.M, Weis, K. | | Deposit date: | 2010-10-27 | | Release date: | 2011-03-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP(6) in mRNA export.
Nature, 472, 2011
|
|
3HSB
 
 | | Crystal structure of YmaH (Hfq) from Bacillus subtilis in complex with an RNA aptamer | | Descriptor: | Protein hfq, RNA (5'-R(*AP*GP*AP*GP*AP*GP*A)-3') | | Authors: | Baba, S, Someya, T, Kumasaka, T, Kawai, G, Nakamura, K. | | Deposit date: | 2009-06-10 | | Release date: | 2010-06-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Hfq from Bacillus subtilis in complex with SELEX-derived RNA aptamer: insight into RNA-binding properties of bacterial Hfq
Nucleic Acids Res., 40, 2012
|
|
6M7Y
 
 | |
5W1H
 
 | |
