1W4T
 
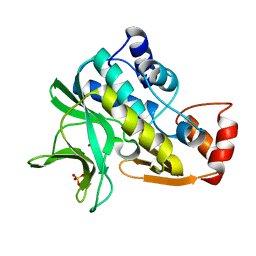 | | X-ray crystallographic structure of Pseudomonas aeruginosa arylamine N-acetyltransferase | | Descriptor: | Arylamine N-acetyltransferase, SULFATE ION | | Authors: | Westwood, I.M, Holton, S.J, Rodrigues-Lima, F, Dupret, J.-M, Bhakta, S, Noble, M.E, Sim, E. | | Deposit date: | 2004-07-29 | | Release date: | 2005-08-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Expression, purification, characterization and structure of Pseudomonas aeruginosa arylamine N-acetyltransferase.
Biochem. J., 385, 2005
|
|
1W2Q
 
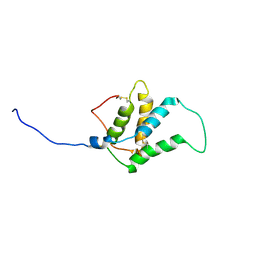 | |
7NEE
 
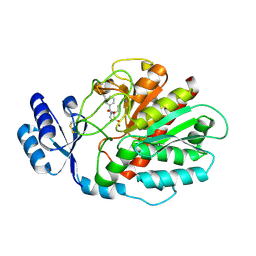 | | Inhibitor Complex with Thrombin Activatable Fibrinolysis inhibitor (TAFIa) | | Descriptor: | (1R,3S)-3-(4-ammoniobutyl)-1-benzyl-1,4-azaphosphinan-1-ium-3-carboxylate 4,4-dioxide, Carboxypeptidase B2, ZINC ION | | Authors: | Brown, D.G, Schaffner, A.P, Gloanec, P, Raimbaud, E, Vuillard, L.M. | | Deposit date: | 2021-02-03 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Phosphinanes and Azaphosphinanes as Potent and Selective Inhibitors of Activated Thrombin-Activatable Fibrinolysis Inhibitor (TAFIa).
J.Med.Chem., 64, 2021
|
|
7NEU
 
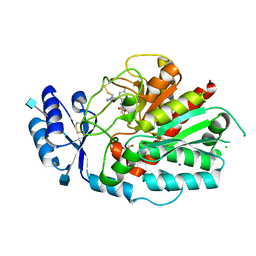 | | Inhibitor Complex with Thrombin Activatable Fibrinolysis Inhibitor (TAFIa) | | Descriptor: | (1R,3S)-3-(4-ammoniobutyl)-1-(4-fluoro-2-(1-methyl-1H-imidazol-5-yl)benzyl)-1,4-azaphosphinan-1-ium-3-carboxylate 4,4-dioxide, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Brown, D.G, Schaffner, A.P, Vuillard, L.M, Gloanec, P, Raimbauld, E. | | Deposit date: | 2021-02-04 | | Release date: | 2021-04-07 | | Last modified: | 2021-04-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Phosphinanes and Azaphosphinanes as Potent and Selective Inhibitors of Activated Thrombin-Activatable Fibrinolysis Inhibitor (TAFIa).
J.Med.Chem., 64, 2021
|
|
1EBW
 
 | | HIV-1 protease in complex with the inhibitor BEA322 | | Descriptor: | HIV-1 PROTEASE, N,N-[2,5-O-[DIBENZYL]-GLUCARYL]-DI-[ISOLEUCYL-AMIDO-METHANE] | | Authors: | Unge, T. | | Deposit date: | 2000-01-25 | | Release date: | 2002-06-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Optimization of P1-P3 groups in symmetric and asymmetric HIV-1 protease inhibitors
Eur.J.Biochem., 270, 2003
|
|
1MMQ
 
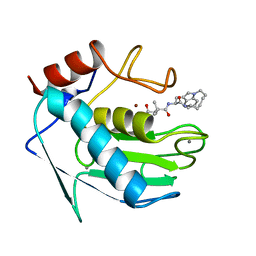 | | MATRILYSIN COMPLEXED WITH HYDROXAMATE INHIBITOR | | Descriptor: | CALCIUM ION, MATRILYSIN, N4-HYDROXY-2-ISOBUTYL-N1-(9-OXO-1,8-DIAZA-TRICYCLO[10.6.1.013,18]NONADECA-12(19),13,15,17-TETRAEN-10-YL)-SUCCINAMIDE, ... | | Authors: | Browner, M.F, Smith, W.W, Castelhano, A.L. | | Deposit date: | 1995-03-22 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Matrilysin-inhibitor complexes: common themes among metalloproteases.
Biochemistry, 34, 1995
|
|
1W7M
 
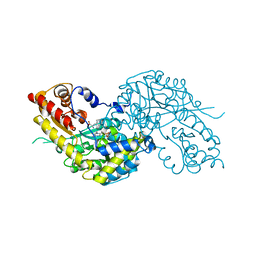 | | Crystal structure of human kynurenine aminotransferase I in complex with L-Phe | | Descriptor: | KYNURENINE--OXOGLUTARATE TRANSAMINASE I, PHENYLALANINE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Rossi, F, Han, Q, Li, J, Li, J, Rizzi, M. | | Deposit date: | 2004-09-06 | | Release date: | 2004-09-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Human Kynurenine Aminotransferase I
J.Biol.Chem., 279, 2004
|
|
1W5Y
 
 | | HIV-1 protease in complex with fluoro substituted diol-based C2- symmetric inhibitor | | Descriptor: | (2R,3R,4R,5R)-2,5-BIS[(2,5-DIFLUOROBENZYL)OXY]-3,4-DIHYDROXY-N,N'-BIS[(1S,2R)-2-HYDROXY-2,3-DIHYDRO-1H-INDEN-1-YL]HEXANEDIAMIDE, POL POLYPROTEIN | | Authors: | Lindberg, J, Pyring, D, Loewgren, S, Rosenquist, A, Zuccarello, G, Kvarnstroem, I, Zhang, H, Vrang, L, Claesson, B, Hallberg, A, Samuelsson, B, Unge, T. | | Deposit date: | 2004-08-10 | | Release date: | 2004-10-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Symmetric Fluoro-Substituted Diol-Based HIV Protease Inhibitors. Ortho-Fluorinated and Meta-Fluorinated P1/P1'-Benzyloxy Side Groups Significantly Improve the Antiviral Activity and Preserve Binding Efficacy
Eur.J.Biochem., 271, 2004
|
|
1W9Z
 
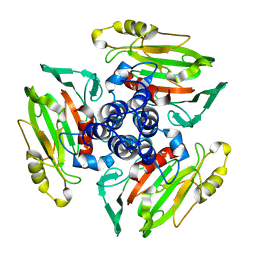 | | Structure of Bannavirus VP9 | | Descriptor: | VP9 | | Authors: | Jaafar, F.M, Attoui, H, Bahar, M.W, Siebold, C, Sutton, G, Mertens, P.P.C, Micco, P, Stuart, D.I, Grimes, J.M, Lamballerie, X. | | Deposit date: | 2004-10-21 | | Release date: | 2005-04-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | The Structure and Function of the Outer Coat Protein Vp9 of Banna Virus
Structure, 13, 2005
|
|
1MR0
 
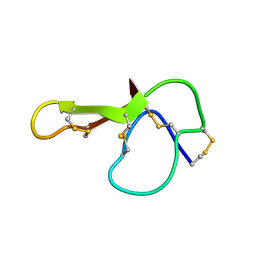 | | SOLUTION NMR STRUCTURE OF AGRP(87-120; C105A) | | Descriptor: | AGOUTI RELATED PROTEIN | | Authors: | Jackson, P.J, Mcnulty, J.C, Yang, Y.K, Thompson, D.A, Chai, B, Gantz, I, Barsh, G.S, Millhauser, G.M. | | Deposit date: | 2002-09-17 | | Release date: | 2002-10-02 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Design, pharmacology, and NMR structure of a minimized cystine knot with agouti-related protein activity.
Biochemistry, 41, 2002
|
|
4PYN
 
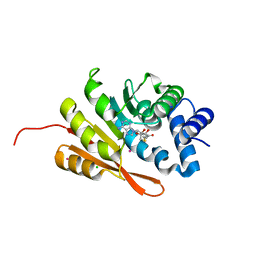 | | Humanized rat COMT in complex with SAH | | Descriptor: | CHLORIDE ION, Catechol O-methyltransferase, POTASSIUM ION, ... | | Authors: | Ehler, A, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2014-03-27 | | Release date: | 2014-06-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Mapping the conformational space accessible to catechol-O-methyltransferase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1ENT
 
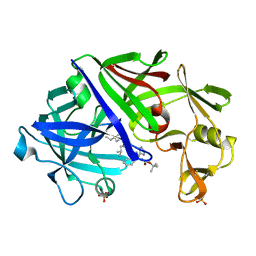 | |
1MTB
 
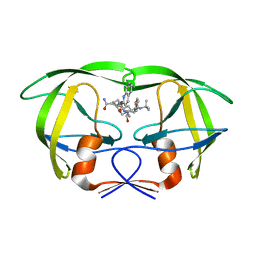 | | Viability of a drug-resistant HIV-1 protease mutant: structural insights for better antiviral therapy | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, PROTEASE RETROPEPSIN | | Authors: | Prabu-Jeyabalan, M, Nalivaika, E.A, King, N.M, Schiffer, C.A. | | Deposit date: | 2002-09-20 | | Release date: | 2003-01-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Viability of drug-resistant human immunodeficiency virus type 1 protease variant: structural insights for better antiviral therapy
J.Virol., 77, 2003
|
|
1MUM
 
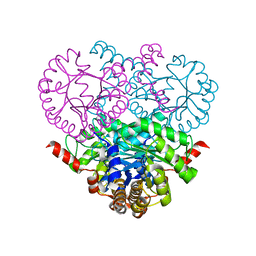 | |
4PY2
 
 | |
1MT7
 
 | | Viability of a drug-resistant HIV-1 protease mutant: structural insights for better antiviral therapy | | Descriptor: | ACETATE ION, PROTEASE RETROPEPSIN, Substrate analogue | | Authors: | Prabu-Jeyabalan, M, Nalivaika, E.A, King, N.M, Schiffer, C.A. | | Deposit date: | 2002-09-20 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Viability of drug-resistant human immunodeficiency virus type 1 protease variant: structural insights for better antiviral therapy
J.Virol., 77, 2003
|
|
8C1Y
 
 | | Small molecule stabilizer for 14-3-3/ChREBP (Cmd 30) | | Descriptor: | 14-3-3 protein sigma, Carbohydrate-responsive element-binding protein, [2-[2-[[2,2-bis(fluoranyl)-2-phenyl-ethyl]amino]-2-oxidanylidene-ethoxy]phenyl]phosphonic acid | | Authors: | Pennings, M.A.M, Visser, E.J, Ottmann, C. | | Deposit date: | 2022-12-21 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular glues of the regulatory ChREBP/14-3-3 complex protect beta cells from glucolipotoxicity.
Biorxiv, 2024
|
|
4PYL
 
 | | Humanized rat COMT in complex with sinefungin, Mg2+, and tolcapone | | Descriptor: | Catechol O-methyltransferase, MAGNESIUM ION, SINEFUNGIN, ... | | Authors: | Ehler, A, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2014-03-27 | | Release date: | 2014-06-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mapping the conformational space accessible to catechol-O-methyltransferase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1EC2
 
 | | HIV-1 protease in complex with the inhibitor BEA428 | | Descriptor: | HIV-1 PROTEASE, N,N-[2,5-O-[DI-4-PYRIDIN-3-YL-BENZYL]-GLUCARYL]-DI-[VALYL-AMIDO-METHANE] | | Authors: | Unge, T. | | Deposit date: | 2000-01-25 | | Release date: | 2002-06-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Optimization of P1-P3 groups in symmetric and asymmetric HIV-1 protease inhibitors
Eur.J.Biochem., 270, 2003
|
|
1MXP
 
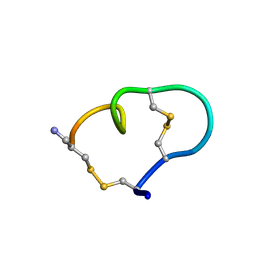 | | Solution structure of the ribbon disulfide bond isomer of alpha-conotoxin AuIB | | Descriptor: | alpha-conotoxin AuIB | | Authors: | Dutton, J.L, Bansal, P.S, Hogg, R.C, Adams, D.J, Alewood, P.F, Craik, D.J. | | Deposit date: | 2002-10-03 | | Release date: | 2002-12-30 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | A New Level of Conotoxin Diversity, a Non-native Disulfide Bond Connectivity in alpha -Conotoxin AuIB Reduces Structural Definition but Increases Biological Activity.
J.Biol.Chem., 277, 2002
|
|
7NPC
 
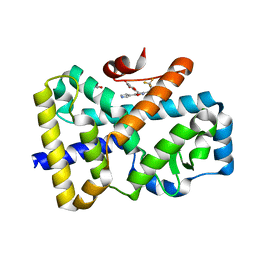 | | ROR(gamma)t ligand binding domain in complex with allosteric ligand FM156 | | Descriptor: | 4-[[3-[2-chloranyl-6-(trifluoromethyl)phenyl]-5-(1~{H}-pyrrol-3-yl)-1,2-oxazol-4-yl]methoxy]benzoic acid, GLYCEROL, Nuclear receptor ROR-gamma | | Authors: | de Vries, R.M.J.M, Meijer, F.A, Brunsveld, L. | | Deposit date: | 2021-02-26 | | Release date: | 2021-06-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structure-Activity Relationship Studies of Trisubstituted Isoxazoles as Selective Allosteric Ligands for the Retinoic-Acid-Receptor-Related Orphan Receptor gamma t.
J.Med.Chem., 64, 2021
|
|
7NP5
 
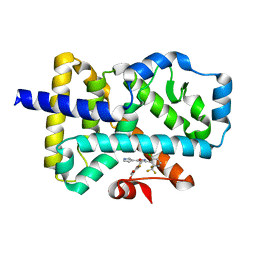 | | ROR(gamma)t ligand binding domain in complex with allosteric ligand FM216 | | Descriptor: | 4-[[3-[2-chloranyl-6-(trifluoromethyl)phenyl]-5-(1~{H}-pyrrol-3-yl)-1,2-oxazol-4-yl]methoxy]-2-fluoranyl-benzoic acid, Nuclear receptor ROR-gamma | | Authors: | Oerlemans, G.J.M, Somsen, B.A, de Vries, R.M.J.M, Meijer, F.A, Brunsveld, L. | | Deposit date: | 2021-02-26 | | Release date: | 2021-06-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-Activity Relationship Studies of Trisubstituted Isoxazoles as Selective Allosteric Ligands for the Retinoic-Acid-Receptor-Related Orphan Receptor gamma t.
J.Med.Chem., 64, 2021
|
|
8INZ
 
 | | Cryo-EM structure of human HCN3 channel in apo state | | Descriptor: | 4-[[(2~{S},4~{a}~{R},6~{S},8~{a}~{S})-6-[(4~{S},5~{R})-4-[(2~{S})-butan-2-yl]-5,9-dimethyl-decyl]-4~{a}-methyl-2,3,4,5,6,7,8,8~{a}-octahydro-1~{H}-naphthalen-2-yl]oxy]-4-oxidanylidene-butanoic acid, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3 | | Authors: | Yu, B, Lu, Q.Y, Li, J, Zhang, J. | | Deposit date: | 2023-03-10 | | Release date: | 2024-04-10 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Cryo-EM structure of human HCN3 channel and its regulation by cAMP.
J.Biol.Chem., 300, 2024
|
|
1W19
 
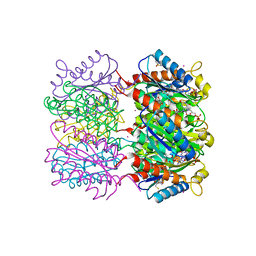 | | Lumazine Synthase from Mycobacterium tuberculosis bound to 3-(1,3,7- trihydro-9-D-ribityl-2,6,8-purinetrione-7-yl)propane 1-phosphate | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, (4S,5S)-1,2-DITHIANE-4,5-DIOL, ... | | Authors: | Morgunova, E, Meining, W, Illarionov, B, Haase, I, Fischer, M, Cushman, M, Bacher, A, Ladenstein, R. | | Deposit date: | 2004-06-03 | | Release date: | 2005-03-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Lumazine Synthase from Mycobacterium Tuberculosis as a Target for Rational Drug Design: Binding Mode of a New Class of Purinetrione Inhibitors(,)
Biochemistry, 44, 2005
|
|
7NEC
 
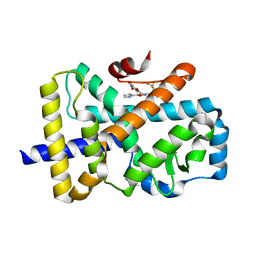 | | ROR(gamma)t ligand binding domain in complex with allosteric ligand FM217 | | Descriptor: | 4-[[3-[2-chloranyl-6-(trifluoromethyl)phenyl]-5-(1~{H}-pyrrol-2-yl)-1,2-oxazol-4-yl]methoxy]benzoic acid, Nuclear receptor ROR-gamma | | Authors: | Somsen, B.A, de Vries, R.M.J.M, Meijer, F.A, Brunsveld, L. | | Deposit date: | 2021-02-03 | | Release date: | 2021-06-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-Activity Relationship Studies of Trisubstituted Isoxazoles as Selective Allosteric Ligands for the Retinoic-Acid-Receptor-Related Orphan Receptor gamma t.
J.Med.Chem., 64, 2021
|
|
