7TTS
 
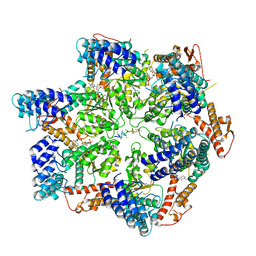 | | Skd3, hexamer, filtered | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Beta-casein, Caseinolytic peptidase B protein homolog, ... | | Authors: | Rizo, A.N, Cupo, R.R. | | Deposit date: | 2022-02-01 | | Release date: | 2022-09-28 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Unique structural features govern the activity of a human mitochondrial AAA+ disaggregase, Skd3.
Cell Rep, 40, 2022
|
|
2ZI2
 
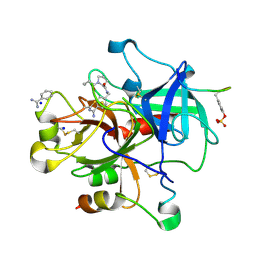 | | Thrombin Inhibition | | Descriptor: | 1-butanoyl-N-(4-carbamimidoylbenzyl)-L-prolinamide, BENZAMIDINE, Hirudin variant-1, ... | | Authors: | Baum, B, Heine, A, Klebe, G. | | Deposit date: | 2008-02-13 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Non-additivity of functional group contributions in protein-ligand binding: a comprehensive study by crystallography and isothermal titration calorimetry.
J.Mol.Biol., 397, 2010
|
|
7PJF
 
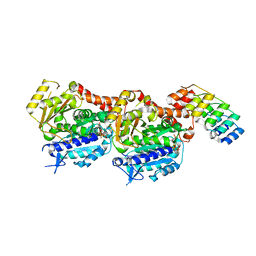 | | Inhibiting parasite proliferation using a rationally designed anti-tubulin agent | | Descriptor: | Designed ankyrin repeat protein (DARPIN) D1, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Sharma, A, Gaillard, N, Ehrhard, V.A, Steinmetz, M.O. | | Deposit date: | 2021-08-24 | | Release date: | 2021-09-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.862 Å) | | Cite: | Inhibiting parasite proliferation using a rationally designed anti-tubulin agent.
Embo Mol Med, 13, 2021
|
|
7FVJ
 
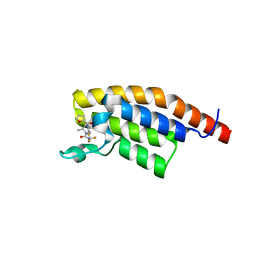 | | PanDDA analysis group deposition -- PHIP in complex with Z4913873236 | | Descriptor: | (2S)-4-(furan-2-carbonyl)-2-methyl-N-(2,2,2-trifluoroethyl)piperazine-1-carboxamide, PH-interacting protein | | Authors: | Grosjean, H, Tomlinson, C, Bradshaw, W.J, Koekemoer, L, Krojer, T, Fearon, D, Biggin, P.C, von Delft, F. | | Deposit date: | 2023-03-09 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7FVA
 
 | | PanDDA analysis group deposition -- PHIP in complex with Z4913872963 | | Descriptor: | (2S)-N-(cyclopropylmethyl)-4-(furan-2-carbonyl)-2-methylpiperazine-1-carboxamide, PH-interacting protein | | Authors: | Grosjean, H, Tomlinson, C, Bradshaw, W.J, Koekemoer, L, Krojer, T, Fearon, D, Biggin, P.C, von Delft, F. | | Deposit date: | 2023-03-09 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7FV9
 
 | | PanDDA analysis group deposition -- PHIP in complex with Z1004253138 | | Descriptor: | 4-(5-methylfuran-2-carbonyl)-N-[(thiophen-2-yl)methyl]piperazine-1-carboxamide, PH-interacting protein | | Authors: | Grosjean, H, Tomlinson, C, Bradshaw, W.J, Koekemoer, L, Krojer, T, Fearon, D, Biggin, P.C, von Delft, F. | | Deposit date: | 2023-03-09 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
2NU9
 
 | |
7PKO
 
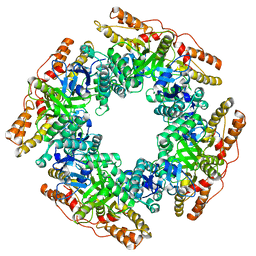 | | CryoEM structure of Rotavirus NSP2 | | Descriptor: | Non-structural protein 2 | | Authors: | Bravo, J.P.K, Borodavka, A. | | Deposit date: | 2021-08-26 | | Release date: | 2021-09-29 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of rotavirus RNA chaperone displacement and RNA annealing.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7FVM
 
 | | PanDDA analysis group deposition -- PHIP in complex with Z1590917771 | | Descriptor: | 4-(furan-2-carbonyl)-N-(1,2-oxazol-3-yl)piperazine-1-carboxamide, PH-interacting protein | | Authors: | Grosjean, H, Tomlinson, C, Bradshaw, W.J, Koekemoer, L, Krojer, T, Fearon, D, Biggin, P.C, von Delft, F. | | Deposit date: | 2023-03-09 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
2NVJ
 
 | | NMR structures of transmembrane segment from subunit a from the yeast proton V-ATPase | | Descriptor: | 25mer peptide from Vacuolar ATP synthase subunit a, vacuolar isoform | | Authors: | Hemminga, M.A, van Mierlo, C.P, Wechselberger, R, de Jong, E.R, Duarte, A.M. | | Deposit date: | 2006-11-13 | | Release date: | 2007-10-02 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Segment TM7 from the cytoplasmic hemi-channel from V(O)-H(+)-V-ATPase includes a flexible region that has a potential role in proton translocation
Biochim.Biophys.Acta, 1768, 2007
|
|
2ZK5
 
 | | Human peroxisome proliferator-activated receptor gamma ligand binding domain complexed with nitro-233 | | Descriptor: | 3-[5-(2-nitropent-1-en-1-yl)furan-2-yl]benzoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Waku, T, Shiraki, T, Oyama, T, Fujimoto, Y, Morikawa, K. | | Deposit date: | 2008-03-12 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural insight into PPARgamma activation through covalent modification with endogenous fatty acids
J.Mol.Biol., 385, 2009
|
|
7FVQ
 
 | | PanDDA analysis group deposition -- PHIP in complex with Z68576046 | | Descriptor: | N-butyl-4-(thiophene-2-carbonyl)piperazine-1-carboxamide, PH-interacting protein | | Authors: | Grosjean, H, Tomlinson, C, Bradshaw, W.J, Koekemoer, L, Krojer, T, Fearon, D, Biggin, P.C, von Delft, F. | | Deposit date: | 2023-03-09 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7FVP
 
 | | PanDDA analysis group deposition -- PHIP in complex with Z1004277578 | | Descriptor: | N-butyl-4-(5-methylfuran-2-carbonyl)piperazine-1-carboxamide, PH-interacting protein | | Authors: | Grosjean, H, Tomlinson, C, Bradshaw, W.J, Koekemoer, L, Krojer, T, Fearon, D, Biggin, P.C, von Delft, F. | | Deposit date: | 2023-03-09 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5LPT
 
 | | tRNA guanine Transglycosylase (TGT) in co-crystallized complex (space group P21) with 6-amino-2-(methylamino)-4-(2-((2R,3R,4S,5R,6S)-3,4,5,6-tetramethoxytetrahydro-2H-pyran-2-yl)ethyl)-1H-imidazo[4,5-g]quinazolin-8(7H)-one | | Descriptor: | 6-azanyl-2-(methylamino)-4-[2-[(2~{R},3~{R},4~{S},5~{R},6~{S})-3,4,5,6-tetramethoxyoxan-2-yl]ethyl]-1,7-dihydroimidazo[4,5-g]quinazolin-8-one, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Ehrmann, F.R, Heine, A, Klebe, G. | | Deposit date: | 2016-08-14 | | Release date: | 2017-08-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Carbohydrate-based Inhibitors targeting the Ribose-34 pocket of Z.mobilis TGT and changing the oligomeric state of the homodimer
To be published
|
|
7FVC
 
 | | PanDDA analysis group deposition -- PHIP in complex with Z495704106 | | Descriptor: | N-(cyclopropylmethyl)-4-(3,4-dichlorobenzoyl)piperazine-1-carboxamide, PH-interacting protein | | Authors: | Grosjean, H, Tomlinson, C, Bradshaw, W.J, Koekemoer, L, Krojer, T, Fearon, D, Biggin, P.C, von Delft, F. | | Deposit date: | 2023-03-09 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
6ZQ6
 
 | |
3I3D
 
 | | E. COLI (lacZ) BETA-GALACTOSIDASE (M542A) IN COMPLEX WITH IPTG | | Descriptor: | 1-methylethyl 1-thio-beta-D-galactopyranoside, Beta-galactosidase, DIMETHYL SULFOXIDE, ... | | Authors: | Dugdale, M.L, Dymianiw, D, Minhas, B, Huber, R.E. | | Deposit date: | 2009-06-30 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Role of Met-542 as a guide for the conformational changes of Phe-601 that occur during the reaction of β-galactosidase (Escherichia coli).
Biochem.Cell Biol., 88, 2010
|
|
7FVN
 
 | | PanDDA analysis group deposition -- PHIP in complex with Z371875396 | | Descriptor: | N-(propan-2-yl)-4-(thiophene-2-carbonyl)piperazine-1-carboxamide, PH-interacting protein | | Authors: | Grosjean, H, Tomlinson, C, Bradshaw, W.J, Koekemoer, L, Krojer, T, Fearon, D, Biggin, P.C, von Delft, F. | | Deposit date: | 2023-03-09 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7FUT
 
 | | PanDDA analysis group deposition -- PHIP in complex with Z44602357 | | Descriptor: | 4-(furan-2-carbonyl)-N-propylpiperazine-1-carboxamide, PH-interacting protein | | Authors: | Grosjean, H, Tomlinson, C, Bradshaw, W.J, Koekemoer, L, Krojer, T, Fearon, D, Biggin, P.C, von Delft, F. | | Deposit date: | 2023-03-09 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5LI6
 
 | | Crystal structure of Mycobacterium tuberculosis CYP126A1 in complex with N-isopropyl-N-((3-(4-methoxyphenyl)-1,2,4-oxadiazol-5-yl)methyl)-2-(4-nitrophenyl)acetamide | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Putative cytochrome P450 126, ~{N}-[[3-(4-methoxyphenyl)-1,2,4-oxadiazol-5-yl]methyl]-2-(4-nitrophenyl)-~{N}-propan-2-yl-ethanamide | | Authors: | Levy, C, Munro, A.W, Leys, D. | | Deposit date: | 2016-07-14 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Characterization and Ligand/Inhibitor Identification Provide Functional Insights into the Mycobacterium tuberculosis Cytochrome P450 CYP126A1.
J. Biol. Chem., 292, 2017
|
|
7FVO
 
 | | PanDDA analysis group deposition -- PHIP in complex with Z1435810807 | | Descriptor: | N-[2-(4,5-dimethyl-1,3-thiazol-2-yl)ethyl]-4-(thiophene-2-carbonyl)piperazine-1-carboxamide, PH-interacting protein | | Authors: | Grosjean, H, Tomlinson, C, Bradshaw, W.J, Koekemoer, L, Krojer, T, Fearon, D, Biggin, P.C, von Delft, F. | | Deposit date: | 2023-03-09 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
6ZS4
 
 | | Crystal structure of the fifth bromodomain of human protein polybromo-1 in complex with tert-butyl 4-[3-amino-6-(2-hydroxyphenyl)pyridazin-4-yl]piperazine-1-carboxylate | | Descriptor: | 1,2-ETHANEDIOL, Protein polybromo-1, tert-butyl 4-[3-amino-6-(2-hydroxyphenyl)pyridazin-4-yl]piperazine-1-carboxylate | | Authors: | Preuss, F, Joerger, A.C, Kraemer, A, Wanior, M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-07-15 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Pan-SMARCA/PB1 Bromodomain Inhibitors and Their Role in Regulating Adipogenesis.
J.Med.Chem., 63, 2020
|
|
2NYL
 
 | | Crystal structure of Protein Phosphatase 2A (PP2A) holoenzyme with the catalytic subunit carboxyl terminus truncated | | Descriptor: | MANGANESE (II) ION, Protein phosphatase 2, regulatory subunit A (PR 65), ... | | Authors: | Xing, Y, Xu, Y, Chen, Y, Chao, Y, Lin, Z, Shi, Y. | | Deposit date: | 2006-11-20 | | Release date: | 2006-12-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure of the Protein Phosphatase 2A Holoenzyme.
Cell(Cambridge,Mass.), 127, 2006
|
|
7FVR
 
 | | PanDDA analysis group deposition -- PHIP in complex with Z166737374 | | Descriptor: | 4-(thiophene-2-carbonyl)-N-[(thiophen-2-yl)methyl]piperazine-1-carboxamide, PH-interacting protein | | Authors: | Grosjean, H, Tomlinson, C, Bradshaw, W.J, Koekemoer, L, Krojer, T, Fearon, D, Biggin, P.C, von Delft, F. | | Deposit date: | 2023-03-09 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7FUV
 
 | | PanDDA analysis group deposition -- PHIP in complex with Z183376720 | | Descriptor: | N-cyclopropyl-4-(furan-2-carbonyl)piperazine-1-carboxamide, PH-interacting protein | | Authors: | Grosjean, H, Tomlinson, C, Bradshaw, W.J, Koekemoer, L, Krojer, T, Fearon, D, Biggin, P.C, von Delft, F. | | Deposit date: | 2023-03-09 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
