7RBN
 
 | | Human DNA polymerase beta crosslinked complex, 20 min Ca to Mg exchange | | Descriptor: | 2-deoxy-3,5-di-O-phosphono-D-erythro-pentitol, DNA (5'-D(*CP*CP*GP*AP*CP*GP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3'), ... | | Authors: | Kumar, A. | | Deposit date: | 2021-07-06 | | Release date: | 2022-03-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Interlocking activities of DNA polymerase beta in the base excision repair pathway.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RBH
 
 | | Human DNA polymerase beta crosslinked ternary complex 2 | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 2-deoxy-3,5-di-O-phosphono-D-erythro-pentitol, CALCIUM ION, ... | | Authors: | Kumar, A. | | Deposit date: | 2021-07-06 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Interlocking activities of DNA polymerase beta in the base excision repair pathway.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RBL
 
 | | Human DNA polymerase beta crosslinked complex, 60 s Ca to Mg exchange | | Descriptor: | 2-deoxy-3,5-di-O-phosphono-D-erythro-pentitol, DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*CP*C)-3'), DNA (5'-D(*GP*TP*CP*GP*G)-3'), ... | | Authors: | Kumar, A. | | Deposit date: | 2021-07-06 | | Release date: | 2022-03-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Interlocking activities of DNA polymerase beta in the base excision repair pathway.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RBF
 
 | | Human DNA polymerase beta crosslinked binary complex - B | | Descriptor: | 1,2-ETHANEDIOL, 2-deoxy-3,5-di-O-phosphono-D-erythro-pentitol, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Reed, A.J, Kumar, A. | | Deposit date: | 2021-07-06 | | Release date: | 2022-03-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Interlocking activities of DNA polymerase beta in the base excision repair pathway.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RBJ
 
 | | Human DNA polymerase beta crosslinked complex, 30 s Ca to Mg exchange | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 2-deoxy-3,5-di-O-phosphono-D-erythro-pentitol, DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*CP*C)-3'), ... | | Authors: | Kumar, A. | | Deposit date: | 2021-07-06 | | Release date: | 2022-03-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Interlocking activities of DNA polymerase beta in the base excision repair pathway.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RBM
 
 | | Human DNA polymerase beta crosslinked complex, 60 s Ca to Mn exchange | | Descriptor: | 2-deoxy-3,5-di-O-phosphono-D-erythro-pentitol, DNA (5'-D(*CP*CP*GP*AP*CP*GP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*CP*C)-3'), ... | | Authors: | Kumar, A. | | Deposit date: | 2021-07-06 | | Release date: | 2022-03-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Interlocking activities of DNA polymerase beta in the base excision repair pathway.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
4ZTZ
 
 | |
5D5X
 
 | | Crystal structure of Chaetomium thermophilum Skn7 with SSRE DNA | | Descriptor: | Putative transcription factor, SSRE DNA strand 1, SSRE DNA strand 2 | | Authors: | Neudegger, T, Verghese, J, Hayer-Hartl, M, Hartl, F.U, Bracher, A. | | Deposit date: | 2015-08-11 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of human heat-shock transcription factor 1 in complex with DNA.
Nat.Struct.Mol.Biol., 23, 2016
|
|
4IHS
 
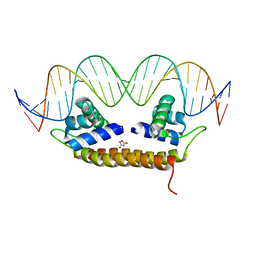 | | Crystal Structure of BenM_DBD/catB site 1 DNA Complex | | Descriptor: | HTH-type transcriptional regulator BenM, MALONATE ION, SODIUM ION, ... | | Authors: | Alanazi, A, Momany, C, Neidle, E.L. | | Deposit date: | 2012-12-19 | | Release date: | 2013-07-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The DNA-binding domain of BenM reveals the structural basis for the recognition of a T-N11-A sequence motif by LysR-type transcriptional regulators.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
8D37
 
 | |
8D33
 
 | |
8D3R
 
 | |
8D42
 
 | |
4IHT
 
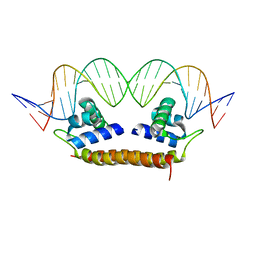 | | Crystal Structure of BenM_DBD/benA site 1 DNA Complex | | Descriptor: | HTH-type transcriptional regulator BenM, benA site 1 DNA, benA site 1 DNA - complement | | Authors: | Alanazi, A, Momany, C, Neidle, E.L. | | Deposit date: | 2012-12-19 | | Release date: | 2013-07-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The DNA-binding domain of BenM reveals the structural basis for the recognition of a T-N11-A sequence motif by LysR-type transcriptional regulators.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
8F6B
 
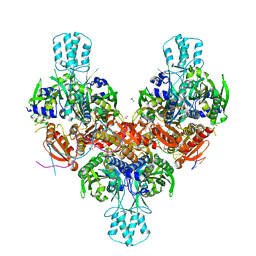 | | Crystal structure of murine PolG2 hexamer bound to DNA | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, DNA (5'-D(*CP*TP*GP*GP*TP*AP*GP*GP*CP*GP*CP*CP*TP*AP*CP*CP*AP*G)-3'), ... | | Authors: | Wojtaszek, J.L, Hoff, K.E, Williams, R.S. | | Deposit date: | 2022-11-16 | | Release date: | 2023-08-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure-specific roles for PolG2-DNA complexes in maintenance and replication of mitochondrial DNA.
Nucleic Acids Res., 51, 2023
|
|
8F69
 
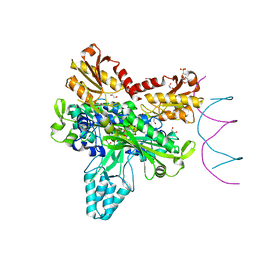 | | Crystal structure of murine PolG2 dimer bound to DNA | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Wojtaszek, J.L, Hoff, K.E, Williams, R.S. | | Deposit date: | 2022-11-16 | | Release date: | 2023-08-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-specific roles for PolG2-DNA complexes in maintenance and replication of mitochondrial DNA.
Nucleic Acids Res., 51, 2023
|
|
5D5W
 
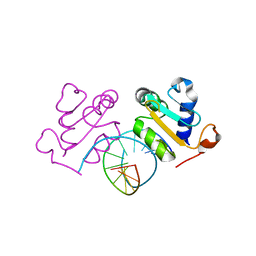 | | Crystal structure of Chaetomium thermophilum Skn7 with HSE DNA | | Descriptor: | HSE DNA, Putative transcription factor | | Authors: | Neudegger, T, Verghese, J, Hayer-Hartl, M, Hartl, F.U, Bracher, A. | | Deposit date: | 2015-08-11 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of human heat-shock transcription factor 1 in complex with DNA.
Nat.Struct.Mol.Biol., 23, 2016
|
|
4WUZ
 
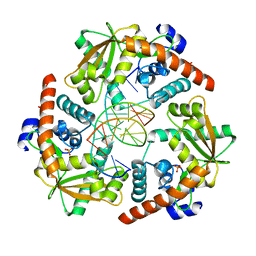 | | Crystal structure of lambda exonuclease in complex with DNA and Ca2+ | | Descriptor: | CALCIUM ION, DNA (5'-D(*TP*T*TP*CP*GP*GP*TP*AP*CP*AP*GP*TP*AP*G)-3'), DNA (5'-D(P*AP*GP*CP*TP*AP*CP*TP*GP*TP*AP*CP*CP*GP*A)-3'), ... | | Authors: | Zhang, J, Bell, C.E. | | Deposit date: | 2014-11-04 | | Release date: | 2014-11-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal Structure of lambda Exonuclease in Complex with DNA and Ca(2+).
Biochemistry, 53, 2014
|
|
2XRO
 
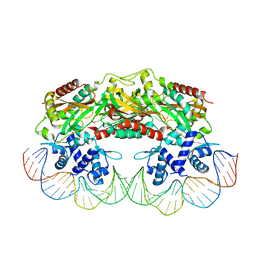 | | Crystal structure of TtgV in complex with its DNA operator | | Descriptor: | HTH-TYPE TRANSCRIPTIONAL REGULATOR TTGV, OSMIUM ION, TTGV OPERATOR DNA | | Authors: | Lu, D, Fillet, S, Meng, C, Alguel, Y, Kloppsteck, P, Bergeron, J, Krell, T, Gallegos, M.-T, Ramos, J, Zhang, X. | | Deposit date: | 2010-09-17 | | Release date: | 2010-12-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal Structure of Ttgv in Complex with its DNA Operator Reveals a General Model for Cooperative DNA Binding of Tetrameric Gene Regulators.
Genes Dev., 24, 2010
|
|
1HLZ
 
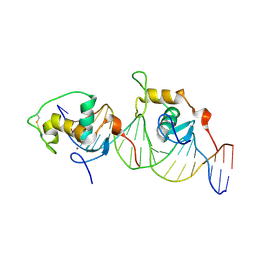 | | CRYSTAL STRUCTURE OF THE ORPHAN NUCLEAR RECEPTOR REV-ERB(ALPHA) DNA-BINDING DOMAIN BOUND TO ITS COGNATE RESPONSE ELEMENT | | Descriptor: | 5'-D(*CP*AP*AP*CP*TP*AP*GP*GP*TP*CP*AP*CP*TP*AP*GP*GP*TP*CP*AP*G)-3', 5'-D(*CP*TP*GP*AP*CP*CP*TP*AP*GP*TP*GP*AP*CP*CP*TP*AP*GP*TP*TP*G)-3', ORPHAN NUCLEAR RECEPTOR NR1D1, ... | | Authors: | Sierk, M.L, Zhao, Q, Rastinejad, F. | | Deposit date: | 2000-12-04 | | Release date: | 2001-11-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | DNA Deformability as a Recognition Feature in the RevErb Response Element
Biochemistry, 40, 2001
|
|
8G9O
 
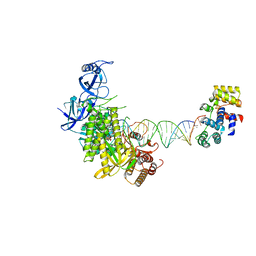 | | Complete DNA elongation subcomplex of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA polymerase alpha catalytic subunit, DNA primase large subunit, ... | | Authors: | Mullins, E.A, Durie, C.L, Ohi, M.D, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-02-21 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
6KRH
 
 | | Structural basis for domain rotation during adenylation of active site K123 and fragment library screening against NAD+ -dependent DNA ligase from Mycobacterium tuberculosis | | Descriptor: | ADENOSINE MONOPHOSPHATE, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, DNA ligase A, ... | | Authors: | Ramachandran, R, Shukla, A, Afsar, M. | | Deposit date: | 2019-08-21 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Salt bridges at the subdomain interfaces of the adenylation domain and active-site residues of Mycobacterium tuberculosis NAD + -dependent DNA ligase A (MtbLigA) are important for the initial steps of nick-sealing activity.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7K04
 
 | | Structure of TFIIH/Rad4-Rad23-Rad33/DNA in DNA opening | | Descriptor: | CALCIUM ION, DNA repair helicase RAD25, DNA repair helicase RAD3, ... | | Authors: | van Eeuwen, T, Min, J.H, Murakami, K. | | Deposit date: | 2020-09-03 | | Release date: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (9.25 Å) | | Cite: | Cryo-EM structure of TFIIH/Rad4-Rad23-Rad33 in damaged DNA opening in nucleotide excision repair.
Nat Commun, 12, 2021
|
|
4TUS
 
 | |
1L9Z
 
 | | Thermus aquaticus RNA Polymerase Holoenzyme/Fork-Junction Promoter DNA Complex at 6.5 A Resolution | | Descriptor: | MAGNESIUM ION, RNA POLYMERASE, ALPHA SUBUNIT, ... | | Authors: | Murakami, K.S, Masuda, S, Campbell, E.A, Muzzin, O, Darst, S.A. | | Deposit date: | 2002-03-27 | | Release date: | 2002-05-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (6.5 Å) | | Cite: | Structural basis of transcription initiation: an RNA polymerase holoenzyme-DNA complex.
Science, 296, 2002
|
|
