1ODS
 
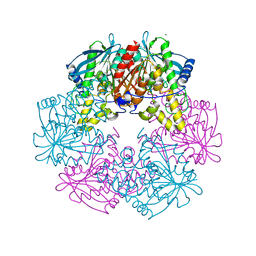 | | Cephalosporin C deacetylase from Bacillus subtilis | | Descriptor: | CEPHALOSPORIN C DEACETYLASE, CHLORIDE ION, MAGNESIUM ION | | Authors: | Vincent, F, Charnock, S.J, Verschueren, K.H.G, Turkenburg, J.P, Scott, D.J, Offen, W.A, Roberts, S, Pell, G, Gilbert, H.J, Brannigan, J.A, Davies, G.J. | | Deposit date: | 2003-02-20 | | Release date: | 2003-07-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Multifunctional Xylooligosaccharide/Cephalosporin C Deacetylase Revealed by the Hexameric Structure of the Bacillus Subtilis Enzyme at 1.9A Resolution
J.Mol.Biol., 330, 2003
|
|
4ARK
 
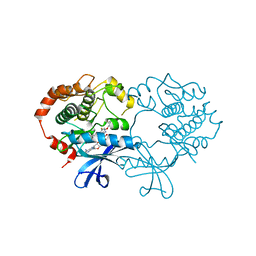 | | CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF HUMAN MAP KINASE KINASE 1 (MEK1) IN COMPLEX WITH A SMALL MOLECULE INHIBITOR AND ADP | | Descriptor: | 2-([3R-3,4-dihydroxybutyl]oxy)-4-fluoro-6-[(2-fluoro-4-iodophenyl)amino]benzamide, ADENOSINE-5'-DIPHOSPHATE, DUAL SPECIFICITY MITOGEN-ACTIVATED PROTEIN KINASE KINASE 1, ... | | Authors: | Hartung, I.V, Hitchcock, M, Puehler, F, Neuhaus, R, Scholz, A, Hammer, S, Petersen, K, Siemeister, G, Brittain, D, Hillig, R.C. | | Deposit date: | 2012-04-24 | | Release date: | 2013-03-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Optimization of Allosteric Mek Inhibitors - Part 1: Venturing Into Unexplored Sar Territories
Bioorg.Med.Chem.Lett., 23, 2013
|
|
2CPZ
 
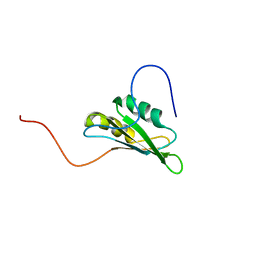 | | solution structure of RNA binding domain 3 in CUG triplet repeat RNA-binding protein 1 | | Descriptor: | CUG triplet repeat RNA-binding protein 1 | | Authors: | Tsuda, K, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-19 | | Release date: | 2005-11-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | solution structure of RNA binding domain 3 in CUG triplet repeat RNA-binding protein 1
To be Published
|
|
1K34
 
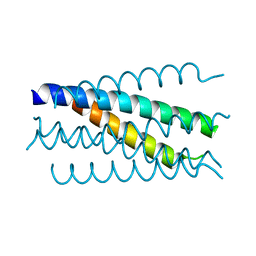 | | Crystal structure analysis of gp41 core mutant | | Descriptor: | Transmembrane glycoprotein GP41 | | Authors: | Shu, W, Lu, M. | | Deposit date: | 2001-10-01 | | Release date: | 2001-10-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Interhelical interactions in the gp41 core: implications for activation of HIV-1 membrane fusion.
Biochemistry, 41, 2002
|
|
3BI7
 
 | | Crystal structure of the SRA domain of E3 ubiquitin-protein ligase UHRF1 | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase UHRF1, SULFATE ION, ... | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Li, Y, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-11-30 | | Release date: | 2007-12-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1.
Nature, 455, 2008
|
|
5SIM
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH C(=O)(N1CCCC1C)c5c(C(Nc3cc2nc(cn2cc3)c4ccccc4)=O)n(nc5)C, micromolar IC50=0.000107 | | Descriptor: | 1-methyl-4-[(2R)-2-methylpyrrolidine-1-carbonyl]-N-[(4R)-2-phenylimidazo[1,2-a]pyridin-7-yl]-1H-pyrazole-5-carboxamide, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Peters, J, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
4DMX
 
 | | Cathepsin K inhibitor | | Descriptor: | (1R,2R)-N-(1-cyanocyclopropyl)-2-{[4-(4-fluorophenyl)piperazin-1-yl]carbonyl}cyclohexanecarboxamide, Cathepsin K, GLYCEROL | | Authors: | Dossetter, A.G, Beeley, H, Bowyer, J, Cook, C.R, Crawford, J.J, Finlayson, J.E, Heron, N.M, Heyes, C, Highton, A.J, Hudson, J.A, Kenny, P.W, Martin, S, MacFaul, P.A, McGuire, T.M, Gutierrez, P.M, Morley, A.D, Morris, J.J, Page, K.M, Rosenbrier Ribeiro, L, Sawney, H, Steinbacher, S, Krapp, S, Jestel, A, Smith, C, Vickers, M. | | Deposit date: | 2012-02-08 | | Release date: | 2012-07-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | (1R,2R)-N-(1-cyanocyclopropyl)-2-(6-methoxy-1,3,4,5-tetrahydropyrido[4,3-b]indole-2-carbonyl)cyclohexanecarboxamide (AZD4996): a potent and highly selective cathepsin K inhibitor for the treatment of osteoarthritis.
J.Med.Chem., 55, 2012
|
|
3ZD2
 
 | | THE STRUCTURE OF THE TWO N-TERMINAL DOMAINS OF COMPLEMENT FACTOR H RELATED PROTEIN 1 SHOWS FORMATION OF A NOVEL DIMERISATION INTERFACE | | Descriptor: | 1,2-ETHANEDIOL, COMPLEMENT FACTOR H-RELATED PROTEIN 1 | | Authors: | Caesar, J.J.E, Goicoechea de Jorge, E, Pickering, M.C, Lea, S.M. | | Deposit date: | 2012-11-23 | | Release date: | 2013-03-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Dimerization of Complement Factor H-Related Proteins Modulates Complement Activation in Vivo.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4DMY
 
 | | Cathepsin K inhibitor | | Descriptor: | (1R,2R)-N-(1-cyanocyclopropyl)-2-[(8-fluoro-1,3,4,5-tetrahydro-2H-pyrido[4,3-b]indol-2-yl)carbonyl]cyclohexanecarboxamide, Cathepsin K, GLYCEROL, ... | | Authors: | Dossetter, A.G, Beeley, H, Bowyer, J, Cook, C.R, Crawford, J.J, Finlayson, J.E, Heron, N.M, Heyes, C, Highton, A.J, Hudson, J.A, Kenny, P.W, Martin, S, MacFaul, P.A, McGuire, T.M, Gutierrez, P.M, Morley, A.D, Morris, J.J, Page, K.M, Rosenbrier Ribeiro, L, Sawney, H, Steinbacher, S, Krapp, S, Jestel, A, Smith, C, Vickers, M. | | Deposit date: | 2012-02-08 | | Release date: | 2012-07-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | (1R,2R)-N-(1-cyanocyclopropyl)-2-(6-methoxy-1,3,4,5-tetrahydropyrido[4,3-b]indole-2-carbonyl)cyclohexanecarboxamide (AZD4996): a potent and highly selective cathepsin K inhibitor for the treatment of osteoarthritis.
J.Med.Chem., 55, 2012
|
|
5Z80
 
 | | Solution structure for the 1:1 complex of a platinum(II)-based tripod bound to a hybrid-1 human telomeric G-quadruplex | | Descriptor: | 4-[1-(2,5,8-triazonia-1$l^4-platinabicyclo[3.3.0]octan-1-yl)pyridin-1-ium-4-yl]-N,N-bis[4-[1-(2,5,8-triazonia-1$l^4-platinabicyclo[3.3.0]octan-1-yl)pyridin-1-ium-4-yl]phenyl]aniline, G-quadruplex DNA (26-MER) | | Authors: | Liu, W.T, Zhong, Y.F, Liu, L.Y, Zeng, W.J, Wang, F.Y, Yang, D.Z, Mao, Z.W. | | Deposit date: | 2018-01-30 | | Release date: | 2018-09-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structures of multiple G-quadruplex complexes induced by a platinum(II)-based tripod reveal dynamic binding
Nat Commun, 9, 2018
|
|
3QI3
 
 | | Crystal structure of PDE9A(Q453E) in complex with inhibitor BAY73-6691 | | Descriptor: | 1-(2-chlorophenyl)-6-[(2R)-3,3,3-trifluoro-2-methylpropyl]-1,7-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, MAGNESIUM ION, ... | | Authors: | Hou, J, Xu, J, Liu, M, Zhao, R, Lou, H, Ke, H. | | Deposit date: | 2011-01-26 | | Release date: | 2011-04-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural asymmetry of phosphodiesterase-9, potential protonation of a glutamic Acid, and role of the invariant glutamine.
Plos One, 6, 2011
|
|
1YAL
 
 | | CARICA PAPAYA CHYMOPAPAIN AT 1.7 ANGSTROMS RESOLUTION | | Descriptor: | CHYMOPAPAIN | | Authors: | Maes, D, Bouckaert, J, Poortmans, F, Wyns, L, Looze, Y. | | Deposit date: | 1996-06-20 | | Release date: | 1996-12-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of chymopapain at 1.7 A resolution.
Biochemistry, 35, 1996
|
|
5SH0
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH C2(N(C)c1cc(ccc1N2C)NC(CN([S](=O)(C)=O)c4c3ccccc3ccc4)=O)=O, micromolar IC50=1.2831377 | | Descriptor: | MAGNESIUM ION, N-(1,3-dimethyl-2-oxo-2,3-dihydro-1H-benzimidazol-5-yl)-N~2~-(methanesulfonyl)-N~2~-naphthalen-1-ylglycinamide, ZINC ION, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Brunner, M, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
3FA6
 
 | |
1PI3
 
 | | E28Q mutant Benzoylformate Decarboxylase From Pseudomonas Putida | | Descriptor: | 2-{3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-4-METHYL-2-OXO-2,3-DIHYDRO-1,3-THIAZOL-5-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, Benzoylformate decarboxylase, CALCIUM ION, ... | | Authors: | Bera, A.K, Hasson, M.S. | | Deposit date: | 2003-05-29 | | Release date: | 2004-11-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | High resolution structure of Benzoylformate Decarboxylate E28Q mutant
To be Published
|
|
4QPA
 
 | |
5RWB
 
 | | INPP5D PanDDA analysis group deposition -- Crystal Structure of the phosphatase and C2 domains of SHIP1 in complex with Z2856434899 | | Descriptor: | 1-[(thiophen-3-yl)methyl]piperidin-4-ol, DIMETHYL SULFOXIDE, Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1 | | Authors: | Bradshaw, W.J, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Regulation of inositol 5-phosphatase activity by the C2 domain of SHIP1 and SHIP2.
Structure, 2024
|
|
5RWU
 
 | | INPP5D PanDDA analysis group deposition -- Crystal Structure of the phosphatase and C2 domains of SHIP1 in complex with Z57778470 | | Descriptor: | 1-methyl-N-[(2-methylphenyl)methyl]-1H-tetrazol-5-amine, DIMETHYL SULFOXIDE, Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1 | | Authors: | Bradshaw, W.J, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Regulation of inositol 5-phosphatase activity by the C2 domain of SHIP1 and SHIP2.
Structure, 2024
|
|
3SWT
 
 | |
4QMS
 
 | | MST3 in complex with DASATINIB | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, N-(2-CHLORO-6-METHYLPHENYL)-2-({6-[4-(2-HYDROXYETHYL)PIPERAZIN-1-YL]-2-METHYLPYRIMIDIN-4-YL}AMINO)-1,3-THIAZOLE-5-CARBOXAMIDE, ... | | Authors: | Olesen, S.H, Watts, C, Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2014-06-16 | | Release date: | 2015-07-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.883 Å) | | Cite: | Discovery of Diverse Small-Molecule Inhibitors of Mammalian Sterile20-like Kinase 3 (MST3).
Chemmedchem, 11, 2016
|
|
4QO9
 
 | | MST3 IN COMPLEX WITH Danusertib | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Olesen, S.H, Watts, C, Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2014-06-19 | | Release date: | 2015-07-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of Diverse Small-Molecule Inhibitors of Mammalian Sterile20-like Kinase 3 (MST3).
Chemmedchem, 11, 2016
|
|
1CBL
 
 | |
1OMD
 
 | | STRUCTURE OF ONCOMODULIN REFINED AT 1.85 ANGSTROMS RESOLUTION. AN EXAMPLE OF EXTENSIVE MOLECULAR AGGREGATION VIA CA2+ | | Descriptor: | CALCIUM ION, ONCOMODULIN | | Authors: | Ahmed, F.R, Przybylska, M, Rose, D.R, Birnbaum, G.I, Pippy, M.E, Macmanus, J.P. | | Deposit date: | 1990-04-19 | | Release date: | 1991-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of oncomodulin refined at 1.85 A resolution. An example of extensive molecular aggregation via Ca2+.
J.Mol.Biol., 216, 1990
|
|
7KPU
 
 | |
1UMS
 
 | | STROMELYSIN-1 CATALYTIC DOMAIN WITH HYDROPHOBIC INHIBITOR BOUND, PH 7.0, 32OC, 20 MM CACL2, 15% ACETONITRILE; NMR ENSEMBLE OF 20 STRUCTURES | | Descriptor: | CALCIUM ION, N-{(2R)-2-[2-(hydroxyamino)-2-oxoethyl]-4-methylpentanoyl}-L-leucyl-L-phenylalaninamide, STROMELYSIN-1, ... | | Authors: | Van Doren, S.R, Kurochkin, A.V, Hu, W, Zuiderweg, E.R.P. | | Deposit date: | 1995-10-31 | | Release date: | 1996-03-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the catalytic domain of human stromelysin complexed with a hydrophobic inhibitor.
Protein Sci., 4, 1995
|
|
