1Q4U
 
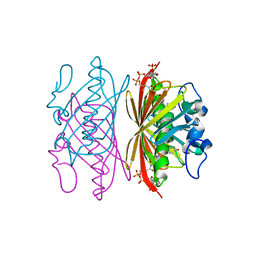 | | Crystal structure of 4-hydroxybenzoyl CoA thioesterase from arthrobacter sp. strain SU complexed with 4-hydroxybenzyl CoA | | Descriptor: | 1,2-ETHANEDIOL, 4-HYDROXYBENZYL COENZYME A, Thioesterase | | Authors: | Thoden, J.B, Zhuang, Z, Dunaway-Mariano, D, Holden, H.M. | | Deposit date: | 2003-08-04 | | Release date: | 2003-09-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Structure of 4-Hydroxybenzoyl-CoA Thioesterase from Arthrobacter sp. strain SU
J.Biol.Chem., 278, 2003
|
|
2OEN
 
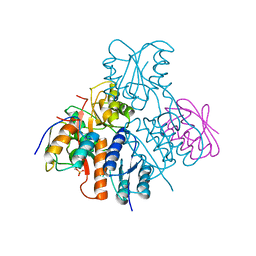 | | Structural mechanism for the fine-tuning of CcpA function by the small molecule effectors glucose-6-phosphate and fructose-1,6-bisphosphate | | Descriptor: | Catabolite control protein, Phosphocarrier protein HPr | | Authors: | Schumacher, M.A, Seidel, G, Hillen, W, Brennan, R.G. | | Deposit date: | 2006-12-30 | | Release date: | 2007-05-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Structural Mechanism for the Fine-tuning of CcpA Function by The Small Molecule Effectors Glucose 6-Phosphate and Fructose 1,6-Bisphosphate.
J.Mol.Biol., 368, 2007
|
|
1FXD
 
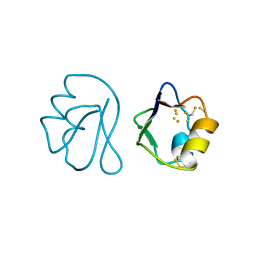 | | REFINED CRYSTAL STRUCTURE OF FERREDOXIN II FROM DESULFOVIBRIO GIGAS AT 1.7 ANGSTROMS | | Descriptor: | FE3-S4 CLUSTER, FERREDOXIN II | | Authors: | Kissinger, C.R, Sieker, L.C, Adman, E.T, Jensen, L.H. | | Deposit date: | 1991-04-08 | | Release date: | 1993-04-15 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Refined crystal structure of ferredoxin II from Desulfovibrio gigas at 1.7 A.
J.Mol.Biol., 219, 1991
|
|
3EBX
 
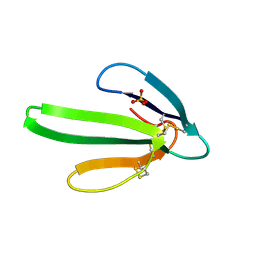 | | REFINEMENT AT 1.4 ANGSTROMS RESOLUTION OF A MODEL OF ERABUTOXIN B. TREATMENT OF ORDERED SOLVENT AND DISCRETE DISORDER | | Descriptor: | ERABUTOXIN B, SULFATE ION | | Authors: | Smith, J.L, Corfield, P.W.R, Hendrickson, W.A, Low, B.W. | | Deposit date: | 1988-01-15 | | Release date: | 1988-04-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Refinement at 1.4 A resolution of a model of erabutoxin b: treatment of ordered solvent and discrete disorder.
Acta Crystallogr.,Sect.A, 44, 1988
|
|
4IGW
 
 | | Crystal structure of kirola (Act d 11) in P6122 space group | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Kirola, ... | | Authors: | Chruszcz, M, Ciardiello, M.A, Giangrieco, I, Osinski, T, Minor, W. | | Deposit date: | 2012-12-18 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural and bioinformatic analysis of the kiwifruit allergen Act d 11, a member of the family of ripening-related proteins.
Mol.Immunol., 56, 2013
|
|
4E32
 
 | | X-ray Structure of the C-3'-Methyltransferase TcaB9 in Complex with S-Adenosyl-L-Homocysteine and dTDP-Sugar Substrate | | Descriptor: | (2R,4S,6R)-4-amino-6-methyl-5-oxotetrahydro-2H-pyran-2-yl [(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)tetrahydrofuran-2-yl]methyl dihydrogen diphosphate, 1,2-ETHANEDIOL, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Bruender, N.A, Holden, H.M. | | Deposit date: | 2012-03-09 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Probing the catalytic mechanism of a C-3'-methyltransferase involved in the biosynthesis of D-tetronitrose.
Protein Sci., 21, 2012
|
|
4IMF
 
 | |
1XII
 
 | |
1XIJ
 
 | |
1G6N
 
 | | 2.1 ANGSTROM STRUCTURE OF CAP-CAMP | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, CATABOLITE GENE ACTIVATOR PROTEIN | | Authors: | Passner, J.M, Schultz, S.C, Steitz, T.A. | | Deposit date: | 2000-11-07 | | Release date: | 2000-12-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Modeling the cAMP-induced allosteric transition using the crystal structure of CAP-cAMP at 2.1 A resolution.
J.Mol.Biol., 304, 2000
|
|
3VT8
 
 | | Crystal structures of rat VDR-LBD with W282R mutation | | Descriptor: | (1R,3R,7E,9beta,17beta)-9-butyl-17-[(2R)-6-hydroxy-6-methylheptan-2-yl]-9,10-secoestra-5,7-diene-1,3-diol, COACTIVATOR PEPTIDE DRIP, Vitamin D3 receptor | | Authors: | Nakabayashi, M, Shimizu, M, Ikura, T, Ito, N. | | Deposit date: | 2012-05-19 | | Release date: | 2013-05-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of hereditary vitamin D-resistant rickets-associated vitamin D receptor mutants R270L and W282R bound to 1,25-dihydroxyvitamin D3 and synthetic ligands.
J.Med.Chem., 56, 2013
|
|
4E31
 
 | | X-ray Structure of the Y76F mutant of TcaB9, a C-3'-Methyltransferase, in Complex with S-Adenosyl-L-Homocysteine and Sugar Product | | Descriptor: | (2R,4S,6R)-4-amino-4,6-dimethyl-5-oxotetrahydro-2H-pyran-2-yl [(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)tetrahydrofuran-2-yl]methyl dihydrogen diphosphate (non-preferred name), 1,2-ETHANEDIOL, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Bruender, N.A, Holden, H.M. | | Deposit date: | 2012-03-09 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Probing the catalytic mechanism of a C-3'-methyltransferase involved in the biosynthesis of D-tetronitrose.
Protein Sci., 21, 2012
|
|
3VUX
 
 | | Crystal structure of A20 ZF7 in complex with linear ubiquitin, form II | | Descriptor: | 1,2-ETHANEDIOL, POTASSIUM ION, Polyubiquitin-C, ... | | Authors: | Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2012-07-09 | | Release date: | 2013-02-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Specific recognition of linear polyubiquitin by A20 zinc finger 7 is involved in NF-kappaB regulation
Embo J., 31, 2012
|
|
4MLM
 
 | | Crystal Structure of PhnZ from uncultured bacterium HF130_AEPn_1 | | Descriptor: | FE (III) ION, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | van Staalduinen, L.M, McSorley, F.R, Zechel, D.L, Jia, Z, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2013-09-06 | | Release date: | 2014-04-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of PhnZ in complex with substrate reveals a di-iron oxygenase mechanism for catabolism of organophosphonates.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4MLY
 
 | | Disulfide isomerase from multidrug resistance IncA/C related integrative and conjugative elements in oxidized state (P21 space group) | | Descriptor: | 1,3-BUTANEDIOL, DsbP | | Authors: | Premkumar, L, Kurth, F, Neyer, S, Martin, J.L. | | Deposit date: | 2013-09-06 | | Release date: | 2013-12-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.207 Å) | | Cite: | The Multidrug Resistance IncA/C Transferable Plasmid Encodes a Novel Domain-swapped Dimeric Protein-disulfide Isomerase.
J.Biol.Chem., 289, 2014
|
|
1Q4T
 
 | | crystal structure of 4-hydroxybenzoyl CoA thioesterase from Arthrobacter sp. strain SU complexed with 4-hydroxyphenyl CoA | | Descriptor: | 1,2-ETHANEDIOL, 4-HYDROXYPHENACYL COENZYME A, CHLORIDE ION, ... | | Authors: | Thoden, J.B, Zhuang, Z, Dunaway-Mariano, D, Holden, H.M. | | Deposit date: | 2003-08-04 | | Release date: | 2003-09-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Structure of 4-Hydroxybenzoyl-CoA Thioesterase from Arthrobacter sp. strain SU
J.Biol.Chem., 278, 2003
|
|
1XRB
 
 | | S-adenosylmethionine synthetase (MAT, ATP: L-methionine S-adenosyltransferase, E.C.2.5.1.6) in which MET residues are replaced with selenomethionine residues (MSE) | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Takusagawa, F, Kamitori, S, Misaki, S, Markham, G.D. | | Deposit date: | 1995-10-26 | | Release date: | 1996-03-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of S-adenosylmethionine synthetase.
J.Biol.Chem., 271, 1996
|
|
3SB4
 
 | |
3D5P
 
 | |
4RE8
 
 | | Crystal Structure of TR3 LBD in complex with Molecule 5 | | Descriptor: | 1-(3,4,5-trihydroxyphenyl)dodecan-1-one, GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Li, F.W, Cai, Q.X, Li, A.Z, Tian, X.Y, Weijia, W, Yuan, W, Hou, P.P, Wu, Q, Lin, T.W. | | Deposit date: | 2014-09-22 | | Release date: | 2015-09-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Induction of Autophagic Death in Cancer Cells by Agonizing TR3 and Attenuating Akt2 Activity
Chem.Biol., 22, 2015
|
|
2O9C
 
 | | Crystal Structure of Bacteriophytochrome chromophore binding domain at 1.45 angstrom resolution | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome | | Authors: | Wagner, J.R, Brunzelle, J.S, Vierstra, R.D, Forest, K.T. | | Deposit date: | 2006-12-13 | | Release date: | 2007-03-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | High resolution structure of deinococcus bacteriophytochrome yields new insights into phytochrome architecture and evolution.
J.Biol.Chem., 282, 2007
|
|
7LV7
 
 | |
7LYH
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with inhibitor UAWJ9-36-1 | | Descriptor: | 3C-like proteinase, GLYCEROL, benzyl (1S,3aR,6aS)-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}carbamoyl)hexahydrocyclopenta[c]pyrrole-2(1H)-carboxylate | | Authors: | Sacco, M, Wang, J, Chen, Y. | | Deposit date: | 2021-03-07 | | Release date: | 2021-03-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rational Design of Hybrid SARS-CoV-2 Main Protease Inhibitors Guided by the Superimposed Cocrystal Structures with the Peptidomimetic Inhibitors GC-376, Telaprevir, and Boceprevir.
Acs Pharmacol Transl Sci, 4, 2021
|
|
1T46
 
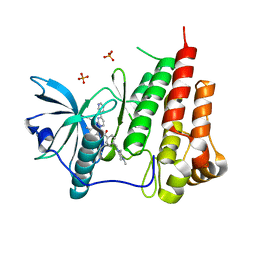 | | STRUCTURAL BASIS FOR THE AUTOINHIBITION AND STI-571 INHIBITION OF C-KIT TYROSINE KINASE | | Descriptor: | 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE, Homo sapiens v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog, PHOSPHATE ION | | Authors: | Mol, C.D, Dougan, D.R, Schneider, T.R, Skene, R.J, Kraus, M.L, Scheibe, D.N, Snell, G.P, Zou, H, Sang, B.C, Wilson, K.P. | | Deposit date: | 2004-04-28 | | Release date: | 2004-06-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the autoinhibition and STI-571 inhibition of c-Kit tyrosine kinase.
J.Biol.Chem., 279, 2004
|
|
7M17
 
 | | SN-407-LRRC8A in MSP1E3D1 lipid nanodiscs (Pose-1) | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 7-{[(2S)-2-butyl-6,7-dichloro-2-cyclopentyl-1-oxo-2,3-dihydro-1H-inden-5-yl]oxy}heptanoic acid, Volume-regulated anion channel subunit LRRC8A | | Authors: | Kern, D.M, Gerber, E.E, Brohawn, S.G. | | Deposit date: | 2021-03-12 | | Release date: | 2021-03-24 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Small molecule SWELL1 complex induction improves glycemic control and nonalcoholic fatty liver disease in murine Type 2 diabetes.
Nat Commun, 13, 2022
|
|
