7S4M
 
 | |
7S4J
 
 | | CryoEM structure of Methylococcus capsulatus (Bath) pMMO in a native lipid nanodisc at 2.16 Angstrom resolution | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dihexanoyl-sn-glycero-3-phosphocholine, Ammonia monooxygenase/methane monooxygenase, ... | | Authors: | Koo, C.W, Rosenzweig, A.C. | | Deposit date: | 2021-09-09 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.16 Å) | | Cite: | Recovery of particulate methane monooxygenase structure and activity in a lipid bilayer.
Science, 375, 2022
|
|
7S4L
 
 | | CryoEM structure of Methylotuvimicrobium alcaliphilum 20Z pMMO in a POPC nanodisc at 2.46 Angstrom resolution | | Descriptor: | (S)-2,3-bis(hexanoyloxy)propyl(2-(trimethylammonio)ethyl)phosphate, 1,2-dihexanoyl-sn-glycero-3-phosphocholine, COPPER (II) ION, ... | | Authors: | Koo, C.W, Rosenzweig, A.C. | | Deposit date: | 2021-09-09 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.46 Å) | | Cite: | Recovery of particulate methane monooxygenase structure and activity in a lipid bilayer.
Science, 375, 2022
|
|
7SIP
 
 | | Structure of shaker-IR | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, POTASSIUM ION, Potassium voltage-gated channel protein Shaker | | Authors: | Tan, X, Bae, C, Stix, R, Fernandez, A.I, Huffer, K, Chang, T, Jiang, J, Faraldo-Gomez, J.D, Swartz, K.J. | | Deposit date: | 2021-10-14 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of the Shaker Kv channel and mechanism of slow C-type inactivation.
Sci Adv, 8, 2022
|
|
7S4I
 
 | | CryoEM structure of Methylococcus capsulatus (Bath) pMMO in a native lipid nanodisc at 2.26 Angstrom resolution | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dihexanoyl-sn-glycero-3-phosphocholine, Ammonia monooxygenase/methane monooxygenase, ... | | Authors: | Koo, C.W, Rosenzweig, A.C. | | Deposit date: | 2021-09-09 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.26 Å) | | Cite: | Recovery of particulate methane monooxygenase structure and activity in a lipid bilayer.
Science, 375, 2022
|
|
7S4H
 
 | | CryoEM structure of Methylococcus capsulatus (Bath) pMMO in a native lipid nanodisc at 2.14 Angstrom resolution | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dihexanoyl-sn-glycero-3-phosphocholine, Ammonia monooxygenase/methane monooxygenase, ... | | Authors: | Koo, C.W, Rosenzweig, A.C. | | Deposit date: | 2021-09-08 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.14 Å) | | Cite: | Recovery of particulate methane monooxygenase structure and activity in a lipid bilayer.
Science, 375, 2022
|
|
7SBX
 
 | | Structure of OC43 spike in complex with polyclonal Fab6 (Donor 1051) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Human polyclonal Fab model with polyalanine backbone - Heavy chain, ... | | Authors: | Bangaru, S, Antanasijevic, A, Ward, A. | | Deposit date: | 2021-09-26 | | Release date: | 2022-05-04 | | Last modified: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural mapping of antibody landscapes to human betacoronavirus spike proteins.
Sci Adv, 8, 2022
|
|
7SBY
 
 | | Structure of OC43 spike in complex with polyclonal Fab7 (Donor 269) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Human polyclonal Fab model with polyalanine backbone - Heavy chain, ... | | Authors: | Bangaru, S, Antanasijevic, A, Ward, A. | | Deposit date: | 2021-09-26 | | Release date: | 2022-05-04 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural mapping of antibody landscapes to human betacoronavirus spike proteins.
Sci Adv, 8, 2022
|
|
7SB3
 
 | | Structure of OC43 spike in complex with polyclonal Fab1 (Donor 269) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Human polyclonal Fab model with polyalanine backbone - Heavy chain, ... | | Authors: | Ward, A, Bangaru, S, Antanasijevic, A. | | Deposit date: | 2021-09-23 | | Release date: | 2022-05-04 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural mapping of antibody landscapes to human betacoronavirus spike proteins.
Sci Adv, 8, 2022
|
|
7SBW
 
 | | Structure of OC43 spike in complex with polyclonal Fab5 (Donor 1051) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Human polyclonal Fab model with polyalanine backbone - Heavy chain, ... | | Authors: | Bangaru, S, Antanasijevic, A, Ward, A. | | Deposit date: | 2021-09-26 | | Release date: | 2022-05-04 | | Last modified: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural mapping of antibody landscapes to human betacoronavirus spike proteins.
Sci Adv, 8, 2022
|
|
7SB4
 
 | | Structure of OC43 spike in complex with polyclonal Fab2 (Donor 1412) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Human polyclonal Fab model with polyalanine backbone - Heavy chain, ... | | Authors: | Bangaru, S, Antanasijevic, A, Ward, A. | | Deposit date: | 2021-09-23 | | Release date: | 2022-05-04 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structural mapping of antibody landscapes to human betacoronavirus spike proteins.
Sci Adv, 8, 2022
|
|
7SB5
 
 | | Structure of OC43 spike in complex with polyclonal Fab3 (Donor 1412) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Human polyclonal Fab model with polyalanine backbone - Heavy chain, ... | | Authors: | Bangaru, S, Antanasijevic, A, Ward, A. | | Deposit date: | 2021-09-23 | | Release date: | 2022-05-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural mapping of antibody landscapes to human betacoronavirus spike proteins.
Sci Adv, 8, 2022
|
|
7SBV
 
 | | Structure of OC43 spike in complex with polyclonal Fab4 (Donor 269) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Human polyclonal Fab model with polyalanine backbone - Heavy chain, ... | | Authors: | Bangaru, S, Antanasijevic, A, Ward, A. | | Deposit date: | 2021-09-26 | | Release date: | 2022-05-04 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural mapping of antibody landscapes to human betacoronavirus spike proteins.
Sci Adv, 8, 2022
|
|
8TRL
 
 | | T cell recognition of citrullinated alpha-enolase peptide presented by HLA-DR4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lim, J.J, Loh, T.J, Reid, H.H, Rossjohn, J. | | Deposit date: | 2023-08-09 | | Release date: | 2024-07-31 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The molecular basis underlying T cell specificity towards citrullinated epitopes presented by HLA-DR4.
Nat Commun, 15, 2024
|
|
7S6I
 
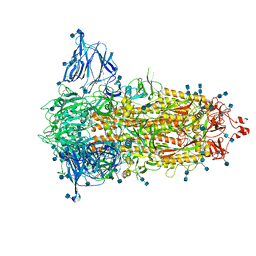 | | SARS-CoV-2-6P-Mut2 S protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Ozorowski, G, Torres, J.L, Ward, A.B. | | Deposit date: | 2021-09-14 | | Release date: | 2022-05-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights of a highly potent pan-neutralizing SARS-CoV-2 human monoclonal antibody.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8TJR
 
 | | CRYO-EM STRUCTURE OF HIV-1 BG505DS-SOSIP.664 ENV TRIMER BOUND TO HERH-c.01 FAB | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody HERH-a.01 Heavy Chain, ... | | Authors: | Morano, N.C, Hoyt, F, Hansen, B, Fischer, E, Shapiro, L. | | Deposit date: | 2023-07-24 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | CRYO-EM STRUCTURE OF HIV-1 BG505DS-SOSIP.664 ENV TRIMER BOUND TO HERH-c.01 FAB
To Be Published
|
|
7S6K
 
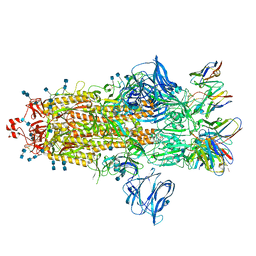 | | J08 fragment antigen binding in complex with SARS-CoV-2-6P-Mut2 S protein (conformation 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, J08 fragment antigen binding heavy chain variable domain, ... | | Authors: | Ozorowski, G, Torres, J.L, Ward, A.B. | | Deposit date: | 2021-09-14 | | Release date: | 2022-05-11 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights of a highly potent pan-neutralizing SARS-CoV-2 human monoclonal antibody.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8TRR
 
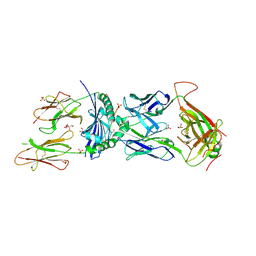 | | T cell recognition of citrullinated vimentin peptide presented by HLA-DR4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Loh, T.J, Lim, J.J, Reid, H.H, Rossjohn, J. | | Deposit date: | 2023-08-10 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The molecular basis underlying T cell specificity towards citrullinated epitopes presented by HLA-DR4.
Nat Commun, 15, 2024
|
|
8TJS
 
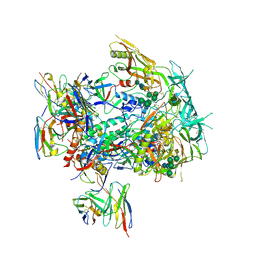 | |
8TNU
 
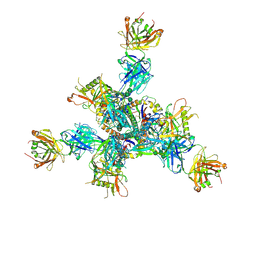 | | Cryo-EM structure of TRNM-b*01 Fab in complex with HIV-1 Env trimer BG505.DS SOSIP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BG505 DS-SOSIP Surface protein gp120, ... | | Authors: | Roark, R.S, Morano, N.C, Shapiro, L.S, Kwong, P.D. | | Deposit date: | 2023-08-02 | | Release date: | 2024-08-07 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Broad and potent HIV-1 neutralization in fusion peptide-primed SHIV-boosted macaques
To Be Published
|
|
8TQ1
 
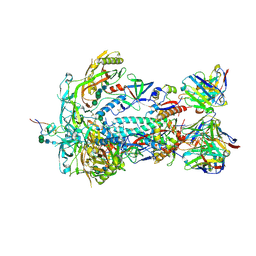 | | HIV-1 BG505 Env SOSIP in complex with bovine Fab Bess4 and non-human primate Fab RM20A3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Bovine Bess4 Fab heavy chain, ... | | Authors: | Ozorowski, G, Lee, W.H, Ward, A.B. | | Deposit date: | 2023-08-06 | | Release date: | 2024-08-07 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Immunization of cows with HIV envelope trimers generates broadly neutralizing antibodies to the V2-apex from the ultralong CDRH3 repertoire.
Plos Pathog., 20, 2024
|
|
8TOP
 
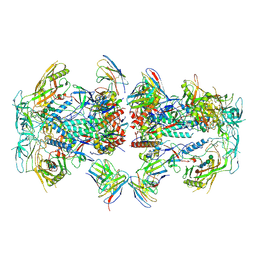 | | Cryo-EM structure of HIV-1 Env BG505 DS-SOSIP in complex with antibody GPZ6-b.01 targeting the fusion peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 BG505 DS-SOSIP glycoprotein gp41, ... | | Authors: | Zhou, T, Morano, N.C, Roark, R.S, Kwong, P.D. | | Deposit date: | 2023-08-03 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Cryo-EM structure of HIV-1 Env BG505 DS-SOSIP in complex with antibody GPZ6-b.01 targeting the fusion peptide
To Be Published
|
|
8TO7
 
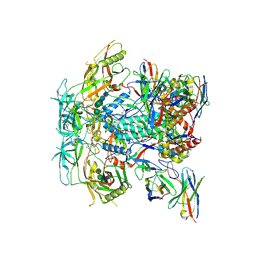 | | Cryo-EM structure of HERH-b*01 Fab in complex with HIV-1 Env trimer BG505.DS SOSIP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HERH-b*01 heavy chain, ... | | Authors: | Roark, R.S, Hoyt, F, Hansen, B, Fischer, E, Shapiro, L.S, Kwong, P.D. | | Deposit date: | 2023-08-03 | | Release date: | 2024-08-07 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Broad and potent HIV-1 neutralization in fusion peptide-primed SHIV-boosted macaques
To Be Published
|
|
8TO9
 
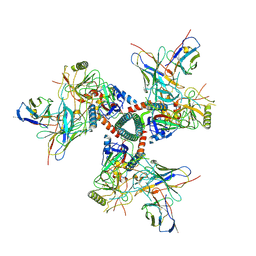 | | Cryo-EM structure of TRNM-f*01 Fab in complex with HIV-1 Env trimer ConC SOSIP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Roark, R.S, Morano, N.C, Shapiro, L.S, Kwong, P.D. | | Deposit date: | 2023-08-03 | | Release date: | 2024-08-07 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.03 Å) | | Cite: | Broad and potent HIV-1 neutralization in fusion peptide-primed SHIV-boosted macaques
To Be Published
|
|
8TPU
 
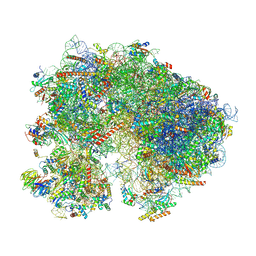 | | Subtomogram averaged consensus structure of the malarial 80S ribosome in Plasmodium falciparum-infected human erythrocytes | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Anton, L, Cheng, W, Zhu, X, Ho, C.M. | | Deposit date: | 2023-08-05 | | Release date: | 2024-08-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Divergent translational landscape reflects adaptation to biased codon usage in malaria parasites
To Be Published
|
|
