2YW3
 
 | |
7DJO
 
 | | The co-crystal structure of DYRK2 with a small molecule inhibitor 17 | | Descriptor: | Dual specificity tyrosine-phosphorylation-regulated kinase 2, [2,7-dimethoxy-9-[[(3S)-pyrrolidin-3-yl]methylsulfanyl]acridin-4-yl]methanol | | Authors: | Wei, T, Xiao, J. | | Deposit date: | 2020-11-20 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Selective inhibition reveals the regulatory function of DYRK2 in protein synthesis and calcium entry.
Elife, 11, 2022
|
|
7POM
 
 | |
7SGA
 
 | | [C-S] DNA mismatch in a self-assembling rhombohedral lattice | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*CP*CP*TP*GP*TP*CP*TP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(P*CP*CP*AP*(IMC)P*AP*CP*A)-3'), DNA (5'-D(P*CP*TP*GP*AP*TP*GP*T)-3'), ... | | Authors: | Vecchioni, S, Lu, B, Bernfeld, W, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2021-10-05 | | Release date: | 2022-10-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Metal-Mediated DNA Nanotechnology in 3D: Structural Library by Templated Diffraction.
Adv Mater, 2023
|
|
2YZC
 
 | | Crystal structure of uricase from Arthrobacter globiformis in complex with allantoate | | Descriptor: | ALLANTOATE ION, Uricase | | Authors: | Juan, E.C.M, Hossain, M.T, Hoque, M.M, Suzuki, K, Sekiguchi, T, Takenaka, A. | | Deposit date: | 2007-05-05 | | Release date: | 2008-05-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Trapping of the uric acid substrate in the crystal structure of urate oxidase from Arthrobacter globiformis
To be Published
|
|
6ZZN
 
 | |
7PX9
 
 | | Substrate-engaged mycobacterial Proteasome-associated ATPase - focused 3D refinement (state A) | | Descriptor: | AAA ATPase forming ring-shaped complexes, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Jomaa, A, Kavalchuk, M, Weber-Ban, E. | | Deposit date: | 2021-10-08 | | Release date: | 2022-01-19 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of prokaryotic ubiquitin-like protein engagement and translocation by the mycobacterial Mpa-proteasome complex.
Nat Commun, 13, 2022
|
|
2LVC
 
 | | Solution NMR Structure of Ig like domain (805-892) of Obscurin-like protein 1 from Homo sapiens, Northeast Structural Genomics Consortium (NESG) Target HR8578K | | Descriptor: | Obscurin-like protein 1 | | Authors: | Pulavarti, S, Eletsky, A, Sukumaran, D.K, Lee, D, Kohan, E, Janjua, H, Xiao, R, Acton, T.B, Everett, J.K, Pederson, K, Prestegard, J, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-06-30 | | Release date: | 2012-08-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of Ig like domain (805-892) of Obscurin-like protein 1 from Homo sapiens, Northeast Structural Genomics Consortium (NESG) Target HR8578K (CASP Target)
To be Published
|
|
2YYK
 
 | | Crystal structure of the mutant of HpaB (T198I, A276G, and R466H) | | Descriptor: | 4-hydroxyphenylacetate-3-hydroxylase, ACETIC ACID, GLYCEROL, ... | | Authors: | Kim, S.-H, Hisano, T, Takeda, K, Iwasaki, W, Ebihara, A, Miki, K. | | Deposit date: | 2007-04-30 | | Release date: | 2007-09-04 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of the Oxygenase Component (HpaB) of the 4-Hydroxyphenylacetate 3-Monooxygenase from Thermus thermophilus HB8
J.Biol.Chem., 282, 2007
|
|
8ON2
 
 | | NI,FE-CODH -600mV state : 24 h Dioxygen Exposure | | Descriptor: | Carbon monoxide dehydrogenase 2, FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Basak, Y, Jeoung, J.-H, Dobbek, H. | | Deposit date: | 2023-03-31 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Stepwise O 2 -Induced Rearrangement and Disassembly of the [NiFe 4 (OH)( mu 3 -S) 4 ] Active Site Cluster of CO Dehydrogenase.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8OMY
 
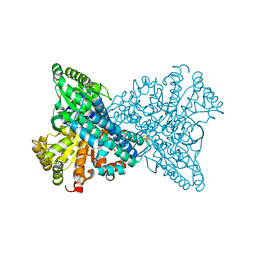 | | NI,FE-CODH -600mV state : 35 min Dioxygen Exposure | | Descriptor: | Carbon monoxide dehydrogenase 2, FE (II) ION, FE (III) ION, ... | | Authors: | Basak, Y, Jeoung, J.-H, Dobbek, H. | | Deposit date: | 2023-03-31 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Stepwise O 2 -Induced Rearrangement and Disassembly of the [NiFe 4 (OH)( mu 3 -S) 4 ] Active Site Cluster of CO Dehydrogenase.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
5K9Q
 
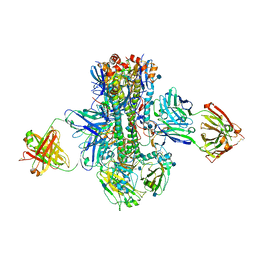 | | Crystal structure of multidonor HV1-18-class broadly neutralizing Influenza A antibody 16.a.26 in complex with A/Hong Kong/1-4-MA21-1/1968 (H3N2) Hemagglutinin | | Descriptor: | 16.a.26 Heavy chain, 16.a.26 Light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Joyce, M.G, Thomas, P.V, Wheatley, A.K, McDermott, A.B, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2016-06-01 | | Release date: | 2016-11-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Vaccine-Induced Antibodies that Neutralize Group 1 and Group 2 Influenza A Viruses.
Cell(Cambridge,Mass.), 166, 2016
|
|
8ON3
 
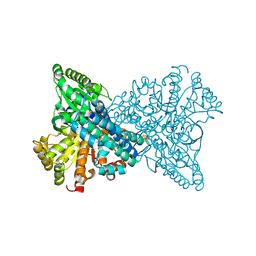 | | NI,FE-CODH -320mV + CN state : 24 h Dioxygen Exposure | | Descriptor: | Carbon monoxide dehydrogenase 2, DI(HYDROXYETHYL)ETHER, FE (II) ION, ... | | Authors: | Basak, Y, Jeoung, J.-H, Dobbek, H. | | Deposit date: | 2023-03-31 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Stepwise O 2 -Induced Rearrangement and Disassembly of the [NiFe 4 (OH)( mu 3 -S) 4 ] Active Site Cluster of CO Dehydrogenase.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7PUH
 
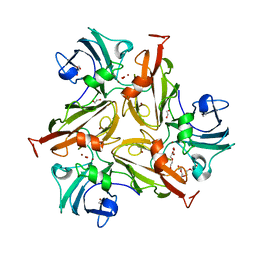 | | Crystal Structure of Two-Domain Laccase mutant H165A/R240H from Streptomyces griseoflavus | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kolyadenko, I, Tishchenko, S, Gabdulkhakov, A. | | Deposit date: | 2021-09-30 | | Release date: | 2022-01-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Engineering the Catalytic Properties of Two-Domain Laccase from Streptomyces griseoflavus Ac-993.
Int J Mol Sci, 23, 2021
|
|
7PXC
 
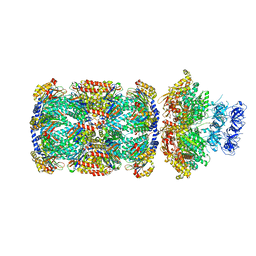 | | Substrate-engaged mycobacterial Proteasome-associated ATPase in complex with open-gate 20S CP - composite map (state A) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Jomaa, A, Kavalchuk, M, Weber-Ban, E. | | Deposit date: | 2021-10-08 | | Release date: | 2022-01-19 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Structural basis of prokaryotic ubiquitin-like protein engagement and translocation by the mycobacterial Mpa-proteasome complex.
Nat Commun, 13, 2022
|
|
8OMX
 
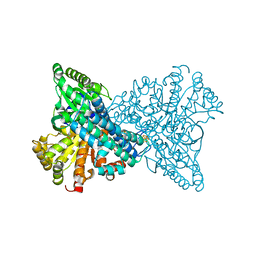 | | NI,FE-CODH -600mV state : 1 min Dioxygen Exposure | | Descriptor: | Carbon monoxide dehydrogenase 2, FE (II) ION, FE(4)-NI(1)-S(5) CLUSTER, ... | | Authors: | Basak, Y, Jeoung, J.-H, Dobbek, H. | | Deposit date: | 2023-03-31 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Stepwise O 2 -Induced Rearrangement and Disassembly of the [NiFe 4 (OH)( mu 3 -S) 4 ] Active Site Cluster of CO Dehydrogenase.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7DQP
 
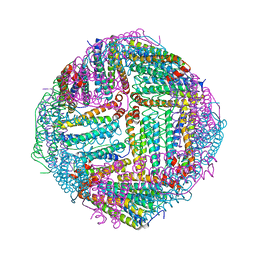 | | Thermal treated Marsupenaeus japonicus ferritin | | Descriptor: | Ferritin | | Authors: | Tan, X, Liu, Y, Zang, J, Zhang, T, Zhao, G. | | Deposit date: | 2020-12-24 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Hyperthermostability of prawn ferritin nanocage facilitates its application as a robust nanovehicle for nutraceuticals.
Int.J.Biol.Macromol., 191, 2021
|
|
8ON1
 
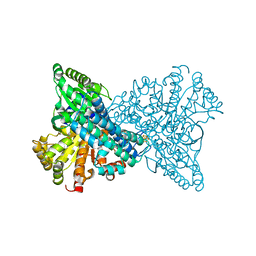 | | NI,FE-CODH -600mV state : 4 h Dioxygen Exposure | | Descriptor: | Carbon monoxide dehydrogenase 2, FE (II) ION, FE (III) ION, ... | | Authors: | Basak, Y, Jeoung, J.-H, Dobbek, H. | | Deposit date: | 2023-03-31 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Stepwise O 2 -Induced Rearrangement and Disassembly of the [NiFe 4 (OH)( mu 3 -S) 4 ] Active Site Cluster of CO Dehydrogenase.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8ON0
 
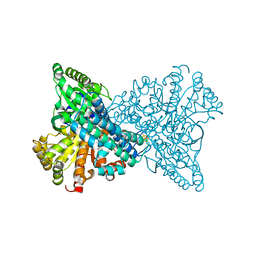 | | NI,FE-CODH -600mV state : 90 min Dioxygen Exposure | | Descriptor: | Carbon monoxide dehydrogenase 2, FE (II) ION, FE (III) ION, ... | | Authors: | Basak, Y, Jeoung, J.-H, Dobbek, H. | | Deposit date: | 2023-03-31 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Stepwise O 2 -Induced Rearrangement and Disassembly of the [NiFe 4 (OH)( mu 3 -S) 4 ] Active Site Cluster of CO Dehydrogenase.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
2LYM
 
 | |
2FR7
 
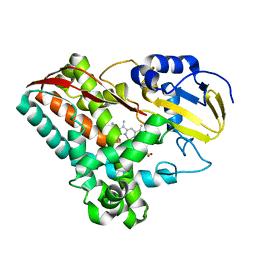 | | Crystal Structure of Cytochrome P450 CYP199A2 | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, putative cytochrome P450 | | Authors: | Rao, Z, Wong, L.L, Xu, F, Bell, S.G. | | Deposit date: | 2006-01-19 | | Release date: | 2007-01-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of CYP199A2, a para-substituted benzoic acid oxidizing cytochrome P450 from Rhodopseudomonas palustris
J.Mol.Biol., 383, 2008
|
|
7PXB
 
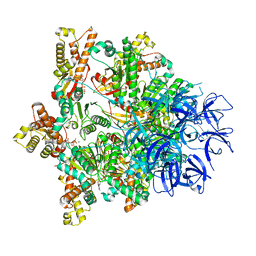 | | Substrate-engaged mycobacterial Proteasome-associated ATPase - focused 3D refinement (state B) | | Descriptor: | AAA ATPase forming ring-shaped complexes, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Jomaa, A, Kavalchuk, M, Weber-Ban, E. | | Deposit date: | 2021-10-08 | | Release date: | 2022-01-19 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of prokaryotic ubiquitin-like protein engagement and translocation by the mycobacterial Mpa-proteasome complex.
Nat Commun, 13, 2022
|
|
7SEP
 
 | |
8P45
 
 | | Crystal structure of human STING in complex with the agonist MD1202D | | Descriptor: | 9-[(1~{S},3~{R},6~{R},8~{R},9~{R},10~{R},12~{R},15~{R},17~{R},18~{R})-8-(6-aminopurin-9-yl)-9,18-bis(fluoranyl)-3,12-bis(oxidanylidene)-3,12-bis(sulfanyl)-2,4,7,11,13-pentaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.2.1.0^{6,10}]octadecan-17-yl]-1~{H}-purin-6-one, Stimulator of interferon genes protein | | Authors: | Klima, M, Boura, E. | | Deposit date: | 2023-05-19 | | Release date: | 2023-12-20 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | Fluorinated cGAMP analogs, which act as STING agonists and are not cleavable by poxins: Structural basis of their function.
Structure, 32, 2024
|
|
8P53
 
 | |
