2J4G
 
 | | Bacteroides thetaiotaomicron GH84 O-GlcNAcase in complex with n-butyl- thiazoline inhibitor | | Descriptor: | (3AR,5R,6S,7R,7AR)-5-(HYDROXYMETHYL)-2-PROPYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D][1,3]THIAZOLE-6,7-DIOL, ACETATE ION, GLYCEROL, ... | | Authors: | Dennis, R.J, Davies, G.J. | | Deposit date: | 2006-08-31 | | Release date: | 2007-01-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Analysis of Pugnac and Nag-Thiazoline as Transition State Analogues for Human O-Glcnacase: Mechanistic and Structural Insights Into Inhibitor Selectivity and Transition State Poise.
J.Am.Chem.Soc., 129, 2007
|
|
4RI8
 
 | |
3JV1
 
 | |
2VO5
 
 | | Structural and biochemical evidence for a boat-like transition state in beta-mannosidases | | Descriptor: | (1R,4R,5R,7R,8R)-2-Benzyl-5-hydroxymethyl-2-aza-bicyclo[2.2.2]octane-4,7,8-triol, 1,2-ETHANEDIOL, BETA-MANNOSIDASE, ... | | Authors: | Tailford, L.E, Offen, W.A, Smith, N.L, Dumon, C, Moreland, C, Gratien, J, Heck, M.P, Stick, R.V, Bleriot, Y, Vasella, A, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2008-02-08 | | Release date: | 2008-04-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Biochemical Evidence for a Boat-Like Transition State in Beta-Mannosidases.
Nat.Chem.Biol., 4, 2008
|
|
4RDW
 
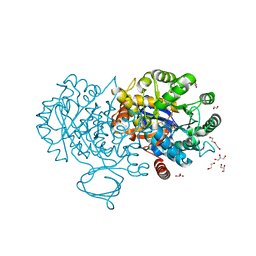 | | The structure of N-formimino-L-Glutamate Iminohydrolase from Pseudomonas aeruginosa complexed with N-Guanidino-L-Glutaric acid | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Marti-Arbona, R, Raushel, F.M, Almo, S.C. | | Deposit date: | 2014-09-19 | | Release date: | 2014-10-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.591 Å) | | Cite: | The structure of N-formimino-L-Glutamate Iminohydrolase from Pseudomonas aeruginosa complexed with N-Guanidino-L-Glutaric acid
To be Published
|
|
4REA
 
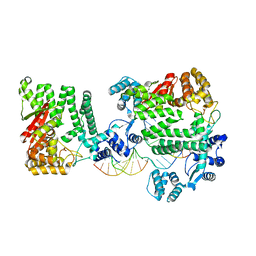 | | A Nuclease DNA complex | | Descriptor: | DNA (5'-D(*TP*GP*CP*TP*CP*GP*CP*CP*AP*C)-3'), DNA (5'-D(P*CP*GP*TP*GP*GP*CP*GP*AP*GP*C)-3'), DNA (5'-D(P*GP*GP*CP*GP*AP*GP*CP*GP*CP*TP*CP*GP*CP*CP*AP*CP*G)-3'), ... | | Authors: | Zhao, Q, Xue, X, Longerich, S, Sung, P, Xiong, Y. | | Deposit date: | 2014-09-22 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.81 Å) | | Cite: | Structural insights into 5' flap DNA unwinding and incision by the human FAN1 dimer.
Nat Commun, 5, 2014
|
|
3JVA
 
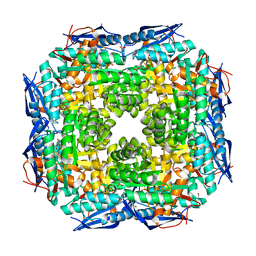 | | Crystal structure of Dipeptide Epimerase from Enterococcus faecalis V583 | | Descriptor: | Dipeptide Epimerase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Sakai, A, Imker, H, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2009-09-16 | | Release date: | 2010-08-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7X5W
 
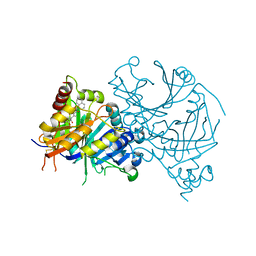 | | Crystal structure of AtHPPD-Y18022 complex | | Descriptor: | 1,5-dimethyl-3-naphthalen-1-yl-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-quinazoline-2,4-dione, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Lin, H.-Y, Dong, J, Yang, G.-F. | | Deposit date: | 2022-03-05 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | Structural insights of 4-Hydrophenylpyruvate dioxygenase inhibition by structurally diverse small molecules
Adv Agrochem, 2022
|
|
3CL6
 
 | | Crystal structure of Puue Allantoinase | | Descriptor: | Puue allantoinase | | Authors: | Ramazzina, I, Cendron, L, Folli, C, Berni, R, Monteverdi, D, Zanotti, G, Percudani, R. | | Deposit date: | 2008-03-18 | | Release date: | 2008-06-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Logical identification of an allantoinase analog (puuE) recruited from polysaccharide deacetylases
J.Biol.Chem., 283, 2008
|
|
7X5Y
 
 | | Crystal structure of AtHPPD-Y14240 complex | | Descriptor: | (1R,3S)-5-methyl-2-[(1S)-2-naphthalen-1-yloxy-1-oxidanyl-ethyl]cyclohexane-1,3-diol, (4S)-2-METHYL-2,4-PENTANEDIOL, 4-hydroxyphenylpyruvate dioxygenase, ... | | Authors: | Lin, H.-Y, Dong, J, Yang, G.-F. | | Deposit date: | 2022-03-05 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.498 Å) | | Cite: | Discovery of Subnanomolar Inhibitors of 4-Hydroxyphenylpyruvate Dioxygenase via Structure-Based Rational Design.
J.Agric.Food Chem., 71, 2023
|
|
7X69
 
 | | Crystal structure of AtHPPD-Y191053 complex | | Descriptor: | 1,5-dimethyl-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-3-(4-phenylbutyl)quinazoline-2,4-dione, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Lin, H.-Y, Dong, J, Yang, G.-F. | | Deposit date: | 2022-03-06 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | Discovery of Subnanomolar Inhibitors of 4-Hydroxyphenylpyruvate Dioxygenase via Structure-Based Rational Design.
J.Agric.Food Chem., 71, 2023
|
|
5WI6
 
 | | Human beta-1 tryptase mutant Ile99Cys | | Descriptor: | L-alpha-glutamyl-N-{(1S)-4-{[amino(iminio)methyl]amino}-1-[(1S)-2-chloro-1-hydroxyethyl]butyl}glycinamide, SULFATE ION, Tryptase alpha/beta-1 | | Authors: | Eigenbrot, C, Maun, H.R. | | Deposit date: | 2017-07-18 | | Release date: | 2018-04-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Dual functionality of beta-tryptase protomers as both proteases and cofactors in the active tetramer.
J. Biol. Chem., 293, 2018
|
|
2J55
 
 | | X-ray reduced Paraccocus denitrificans methylamine dehydrogenase O- quinone in complex with amicyanin. | | Descriptor: | AMICYANIN, COPPER (II) ION, GLYCEROL, ... | | Authors: | Pearson, A.R, Pahl, R, Davidson, V.L, Wilmot, C.M. | | Deposit date: | 2006-09-12 | | Release date: | 2007-01-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Tracking X-Ray-Derived Redox Changes in Crystals of a Methylamine Dehydrogenase/Amicyanin Complex Using Single-Crystal Uv/Vis Microspectrophotometry.
J.Synchrotron Radiat., 14, 2007
|
|
7X8H
 
 | | Crystal structure of AtHPPD-(+)-Usnic acid complex | | Descriptor: | (9bR)-2,6-diethanoyl-8,9b-dimethyl-3,7,9-tris(oxidanyl)dibenzofuran-1-one, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Dong, J, Lin, H.-Y, Yang, G.-F. | | Deposit date: | 2022-03-13 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.986 Å) | | Cite: | Discovery of Subnanomolar Inhibitors of 4-Hydroxyphenylpyruvate Dioxygenase via Structure-Based Rational Design.
J.Agric.Food Chem., 71, 2023
|
|
3CU9
 
 | | High resolution crystal structure of 1,5-alpha-L-arabinanase from Geobacillus Stearothermophilus | | Descriptor: | CALCIUM ION, GLYCEROL, Intracellular arabinanase | | Authors: | Alhassid, A, Ben David, A, Shoham, Y, Shoham, G. | | Deposit date: | 2008-04-16 | | Release date: | 2009-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Crystal structure of an inverting GH 43 1,5-alpha-L-arabinanase from Geobacillus stearothermophilus complexed with its substrate
Biochem.J., 422, 2009
|
|
7X8E
 
 | | Crystal structure of PfHPPD-Y13287 complex | | Descriptor: | 1,5-dimethyl-3-(2-methylphenyl)-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-quinazoline-2,4-dione, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Dong, J, Lin, H.-Y, Yang, G.-F. | | Deposit date: | 2022-03-12 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.749 Å) | | Cite: | Structural insights of 4-Hydrophenylpyruvate dioxygenase inhibition by structurally diverse small molecules
Adv Agrochem, 2022
|
|
2IXA
 
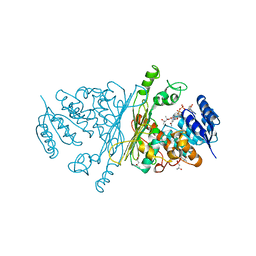 | | A-zyme, N-acetylgalactosaminidase | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ALPHA-N-ACETYLGALACTOSAMINIDASE, ... | | Authors: | Sulzenbacher, G, Liu, Q.P, Bourne, Y, Henrissat, B, Clausen, H. | | Deposit date: | 2006-07-07 | | Release date: | 2007-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bacterial Glycosidases for the Production of Universal Red Blood Cells.
Nat.Biotechnol., 25, 2007
|
|
7X5S
 
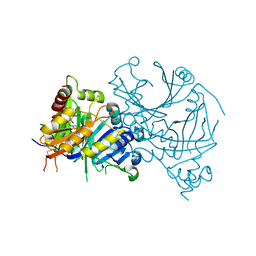 | | Crystal structure of AtHPPD-Y14157 complex | | Descriptor: | 2-methyl-4-(2-nitro-4-piperidin-1-yl-phenyl)carbonyl-5-phenyl-1H-pyrazol-3-one, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Lin, H.-Y, Dong, J, Yang, G.-F. | | Deposit date: | 2022-03-05 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.706 Å) | | Cite: | Structural insights of 4-Hydrophenylpyruvate dioxygenase inhibition by structurally diverse small molecules
Adv Agrochem, 2022
|
|
2J4S
 
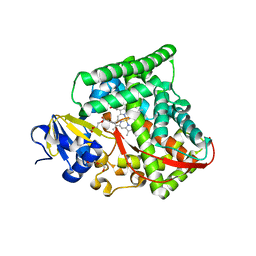 | | P450 BM3 heme domain in complex with DMSO | | Descriptor: | BIFUNCTIONAL P-450:NADPH-P450 REDUCTASE, DI(HYDROXYETHYL)ETHER, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Kuper, J, Tuck-Seng, W, Roccatano, D, Wilmanns, M, Schwaneberg, U. | | Deposit date: | 2006-09-05 | | Release date: | 2007-05-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Understanding a Mechanism of Organic Cosolvent Inactivation in Heme Monooxygenase P450 Bm-3.
J.Am.Chem.Soc., 129, 2007
|
|
7X5X
 
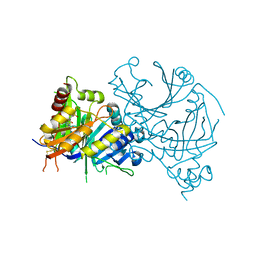 | | Crystal structure of AtHPPD-YH20533 complex | | Descriptor: | 2-methylsulfonyl-N-(1-methyl-1,2,3,4-tetrazol-5-yl)-4-(trifluoromethyl)benzamide, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Lin, H.-Y, Dong, J, Yang, G.-F. | | Deposit date: | 2022-03-05 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.196 Å) | | Cite: | Structural insights of 4-Hydrophenylpyruvate dioxygenase inhibition by structurally diverse small molecules
Adv Agrochem, 2022
|
|
5WIZ
 
 | | Phosphotriesterase variant S5 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, Phosphotriesterase, ... | | Authors: | Miton, C.M, Campbell, E.C, Jackson, C.J, Tokuriki, N. | | Deposit date: | 2017-07-21 | | Release date: | 2019-01-23 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Phosphotriesterase variant S5
To Be Published
|
|
4L3H
 
 | |
7X5Z
 
 | | Crystal structure of AtHPPD-Y13162 complex | | Descriptor: | 1-methyl-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-3-phenyl-quinazoline-2,4-dione, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Lin, H.-Y, Dong, J, Yang, G.-F. | | Deposit date: | 2022-03-05 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.688 Å) | | Cite: | Discovery of Subnanomolar Inhibitors of 4-Hydroxyphenylpyruvate Dioxygenase via Structure-Based Rational Design.
J.Agric.Food Chem., 71, 2023
|
|
5WJA
 
 | | Crystal structure of H107A peptidylglycine alpha-hydroxylating monooxygenase (PHM) in complex with citrate | | Descriptor: | CITRATE ANION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Maheshwari, S, Rudzka, K, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2017-07-21 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Effects of copper occupancy on the conformational landscape of peptidylglycine alpha-hydroxylating monooxygenase.
Commun Biol, 1, 2018
|
|
7X67
 
 | | Crystal structure of AtHPPD-Y18406 complex | | Descriptor: | 1,5-dimethyl-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-3-(3-phenylpropyl)quinazoline-2,4-dione, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Lin, H.-Y, Dong, J, Yang, G.-F. | | Deposit date: | 2022-03-06 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | Discovery of Subnanomolar Inhibitors of 4-Hydroxyphenylpyruvate Dioxygenase via Structure-Based Rational Design.
J.Agric.Food Chem., 71, 2023
|
|
