8STY
 
 | |
8STZ
 
 | |
8T27
 
 | |
8T26
 
 | |
4L1M
 
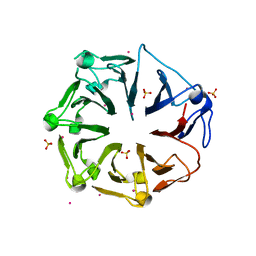 | | Structure of the first RCC1-like domain of HERC2 | | Descriptor: | E3 ubiquitin-protein ligase HERC2, SULFATE ION, UNKNOWN ATOM OR ION | | Authors: | Tempel, W, Khan, M.B, Dong, A, Hu, J, Li, Y, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-06-03 | | Release date: | 2013-07-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the first RCC1-like domain of HERC2
TO BE PUBLISHED
|
|
7VPD
 
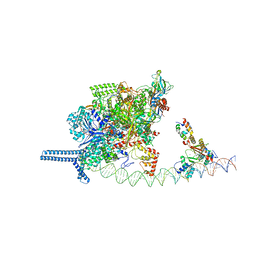 | |
5BTS
 
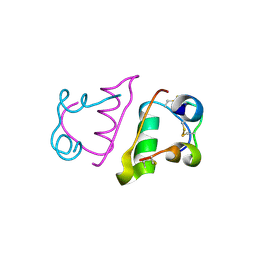 | |
7SDA
 
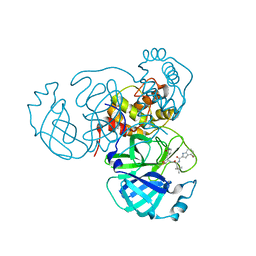 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI49 | | Descriptor: | 3C-like proteinase, N-[(2S)-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-4,4-dimethyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Yang, K.S, Liu, W.R. | | Deposit date: | 2021-09-29 | | Release date: | 2022-11-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Systematic Survey of Reversibly Covalent Dipeptidyl Inhibitors of the SARS-CoV-2 Main Protease.
J.Med.Chem., 66, 2023
|
|
3HH8
 
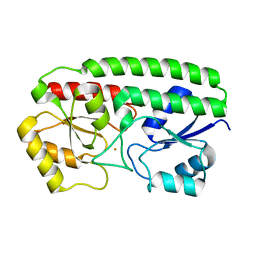 | | Crystal Structure and metal binding properties of the lipoprotein MtsA | | Descriptor: | FE (III) ION, Metal ABC transporter substrate-binding lipoprotein | | Authors: | Baker, E.N, Baker, H.M, Sun, X, Ye, Q.-Y. | | Deposit date: | 2009-05-15 | | Release date: | 2009-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structure and metal binding properties of the lipoprotein MtsA, responsible for iron transport in Streptococcus pyogenes.
Biochemistry, 48, 2009
|
|
7SD9
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI48 | | Descriptor: | 3C-like proteinase, N-[(2S)-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-4,4-dimethyl-1-oxopentan-2-yl]-1H-indole-2-carboxamide | | Authors: | Yang, K.S, Liu, W.R. | | Deposit date: | 2021-09-29 | | Release date: | 2022-11-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Systematic Survey of Reversibly Covalent Dipeptidyl Inhibitors of the SARS-CoV-2 Main Protease.
J.Med.Chem., 66, 2023
|
|
7SDC
 
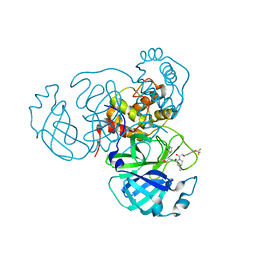 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MI-09 | | Descriptor: | (1R,2S,5S)-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-{[4-(trifluoromethoxy)phenoxy]acetyl}-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Yang, K.S, Liu, W.R. | | Deposit date: | 2021-09-29 | | Release date: | 2022-11-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Systematic Survey of Reversibly Covalent Dipeptidyl Inhibitors of the SARS-CoV-2 Main Protease.
J.Med.Chem., 66, 2023
|
|
8OMZ
 
 | |
1TTV
 
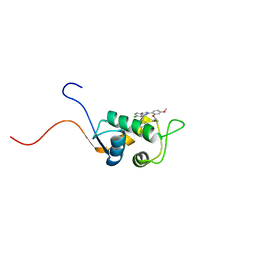 | | NMR Structure of a Complex Between MDM2 and a Small Molecule Inhibitor | | Descriptor: | 1-{[4,5-BIS(4-CHLOROPHENYL)-2-(2-ISOPROPOXY-4-METHOXYPHENYL)-4,5-DIHYDRO-1H-IMIDAZOL-1-YL]CARBONYL}PIPERAZINE, Ubiquitin-protein ligase E3 Mdm2 | | Authors: | Fry, D.C, Emerson, S.D, Palme, S, Vu, B.T, Liu, C.M, Podlaski, F. | | Deposit date: | 2004-06-23 | | Release date: | 2005-01-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a complex between MDM2 and a small molecule inhibitor.
J.Biomol.Nmr, 30, 2004
|
|
1G0Q
 
 | | CRYSTAL STRUCTURE OF T4 LYSOZYME MUTANT V149I | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, PROTEIN (LYSOZYME) | | Authors: | Xu, J, Baase, W.A, Quillin, M.L, Matthews, B.W. | | Deposit date: | 2000-10-06 | | Release date: | 2001-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and thermodynamic analysis of the binding of solvent at internal sites in T4 lysozyme.
Protein Sci., 10, 2001
|
|
2B99
 
 | | Crystal Structure of an archaeal pentameric riboflavin synthase Complex with a Substrate analog inhibitor | | Descriptor: | 6,7-DIOXO-5H-8-RIBITYLAMINOLUMAZINE, Riboflavin synthase | | Authors: | Ramsperger, A, Augustin, M, Schott, A.K, Gerhardt, S, Krojer, T, Eisenreich, W, Illarionov, B, Cushman, M, Bacher, A, Huber, R, Fischer, M. | | Deposit date: | 2005-10-11 | | Release date: | 2005-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal Structure of an Archaeal Pentameric Riboflavin Synthase in Complex with a Substrate Analog Inhibitor: stereochemical implications
J.Biol.Chem., 281, 2006
|
|
2V9T
 
 | |
5BWW
 
 | |
1SVH
 
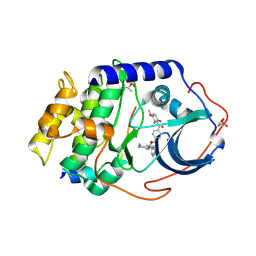 | | Crystal Structure of Protein Kinase A in Complex with Azepane Derivative 8 | | Descriptor: | (3R,4S)-N-(4-{TRANS-2-[4-(2-FLUORO-6-HYDROXY-3-METHOXY-BENZOYL)-PHENYL]-VINYL}-AZEPAN-3-YL)-ISONICOTINAMIDE, cAMP-dependent protein kinase inhibitor, alpha form, ... | | Authors: | Breitenlechner, C.B, Wegge, T, Berillon, L, Graul, K, Marzenell, K, Friebe, W.G, Thomas, U, Schumacher, R, Huber, R, Engh, R.A, Masjost, B. | | Deposit date: | 2004-03-29 | | Release date: | 2005-03-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-based optimization of novel azepane derivatives as PKB inhibitors
J.Med.Chem., 47, 2004
|
|
1T4E
 
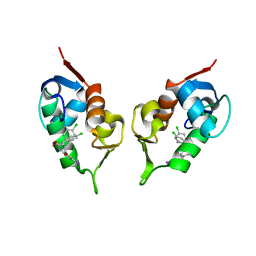 | | Structure of Human MDM2 in complex with a Benzodiazepine Inhibitor | | Descriptor: | (4-CHLOROPHENYL)[3-(4-CHLOROPHENYL)-7-IODO-2,5-DIOXO-1,2,3,5-TETRAHYDRO-4H-1,4-BENZODIAZEPIN-4-YL]ACETIC ACID, Ubiquitin-protein ligase E3 Mdm2 | | Authors: | Grasberger, B.L, Schubert, C, Koblish, H.K, Carver, T.E, Franks, C.F, Zhao, S.Y, Lu, T, LaFrance, L.V, Parks, D.J. | | Deposit date: | 2004-04-29 | | Release date: | 2005-02-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery and cocrystal structure of benzodiazepinedione HDM2 antagonists that activate p53 in cells
J.Med.Chem., 48, 2005
|
|
1IWD
 
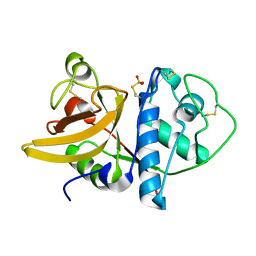 | |
4BTM
 
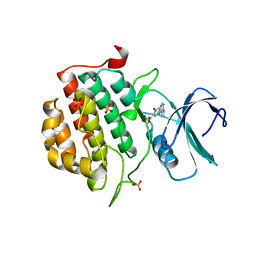 | | TTBK1 in complex with inhibitor | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, SULFATE ION, ... | | Authors: | Xue, Y, Wan, P, Hillertz, P, Schweikart, F, Zhao, Y, Wissler, L, Dekker, N. | | Deposit date: | 2013-06-18 | | Release date: | 2013-09-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | X-Ray Structural Analysis of Tau-Tubulin Kinase 1 and its Interactions with Small Molecular Inhibitors.
Chemmedchem, 8, 2013
|
|
2CCV
 
 | | Structure of Helix Pomatia agglutinin with zinc and N-acetyl-alpha-D- galactoseamine (GalNAc) | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Sanchez, J.-F, Lescar, J, Chazalet, V, Audfray, A, Gautier, C, Gagnon, J, Breton, C, Imberty, A, Mitchell, E.P. | | Deposit date: | 2006-01-18 | | Release date: | 2006-05-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Biochemical and Structural Analysis of Helix Pomatia Agglutinin. A Hexameric Lectin with a Novel Fold.
J.Biol.Chem., 281, 2006
|
|
1YTG
 
 | | SIV PROTEASE CRYSTALLIZED WITH PEPTIDE PRODUCT | | Descriptor: | HIV PROTEASE, PEPTIDE PRODUCT | | Authors: | Rose, R.B, Craik, C.S, Douglas, N.L, Stroud, R.M. | | Deposit date: | 1996-08-01 | | Release date: | 1997-03-12 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Three-dimensional structures of HIV-1 and SIV protease product complexes.
Biochemistry, 35, 1996
|
|
3ZKY
 
 | | Isopenicillin N synthase with substrate analogue AhCmC | | Descriptor: | FE (III) ION, GLYCEROL, ISOPENICILLIN N SYNTHASE, ... | | Authors: | Daruzzaman, A, Clifton, I.J, Rutledge, P.J. | | Deposit date: | 2013-01-25 | | Release date: | 2013-03-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Interaction of Isopenicillin N Synthase with Homologated Substrate Analogues Delta-(L-Alpha-Aminoadipoyl)-L-Homocysteinyl-D-Xaa Characterised by Protein Crystallography.
Chembiochem, 14, 2013
|
|
4JES
 
 | | 1.6A resolution Apo structure of the hemophore HasA from Yersinia pestis (Hexagonal Form) | | Descriptor: | HEXAETHYLENE GLYCOL, Hemophore HasA, MALONATE ION, ... | | Authors: | Kumar, R, Lovell, S, Battaile, K.P, Rivera, M. | | Deposit date: | 2013-02-27 | | Release date: | 2013-04-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Hemophore HasA from Yersinia pestis (HasAyp) Coordinates Hemin with a Single Residue, Tyr75, and with Minimal Conformational Change.
Biochemistry, 52, 2013
|
|
