1SS1
 
 | | STAPHYLOCOCCAL PROTEIN A, B-DOMAIN, Y15W MUTANT, NMR, 25 STRUCTURES | | Descriptor: | Immunoglobulin G binding protein A | | Authors: | Sato, S, Religa, T.L, Daggett, V, Fersht, A.R. | | Deposit date: | 2004-03-23 | | Release date: | 2004-04-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | From The Cover: Testing protein-folding simulations by experiment: B domain of protein A.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
3NDD
 
 | |
3NDF
 
 | |
4UQV
 
 | |
6PCU
 
 | |
3WKJ
 
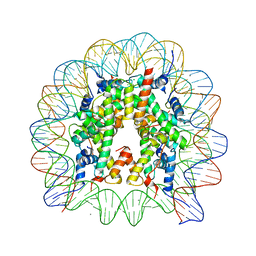 | | The nucleosome containing human TSH2B | | Descriptor: | CHLORIDE ION, DNA (145-MER), Histone H2A type 1-B/E, ... | | Authors: | Urahama, T, Horikoshi, N, Osakabe, A, Tachiwana, H, Kurumizaka, H. | | Deposit date: | 2013-10-22 | | Release date: | 2014-04-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of human nucleosome containing the testis-specific histone variant TSH2B.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
3WUT
 
 | | Structure basis of inactivating cell abscission | | Descriptor: | Centrosomal protein of 55 kDa, GLYCEROL, Inactive serine/threonine-protein kinase TEX14 | | Authors: | Kim, H.J, Matsuura, A, Lee, H.H. | | Deposit date: | 2014-05-05 | | Release date: | 2015-07-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structural and biochemical insights into the role of testis-expressed gene 14 (TEX14) in forming the stable intercellular bridges of germ cells.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3WUV
 
 | | Structure basis of inactivating cell abscission with chimera peptide 2 | | Descriptor: | Centrosomal protein of 55 kDa, peptide from Programmed cell death 6-interacting protein | | Authors: | Kim, H.J, Matsuura, A, Lee, H.H. | | Deposit date: | 2014-05-05 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural and biochemical insights into the role of testis-expressed gene 14 (TEX14) in forming the stable intercellular bridges of germ cells.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3WUU
 
 | |
2GWO
 
 | | crystal structure of TMDP | | Descriptor: | Dual specificity protein phosphatase 13 | | Authors: | Kim, S.J, Ryu, S.E, Kim, J.H. | | Deposit date: | 2006-05-05 | | Release date: | 2007-03-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of human TMDP, a testis-specific dual specificity protein phosphatase: implications for substrate specificity
Proteins, 66, 2007
|
|
2IUL
 
 | | Human tACE g13 mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ANGIOTENSIN-CONVERTING ENZYME, ... | | Authors: | Watermeyer, J.M, Sewell, B.T, Natesh, R, Corradi, H.R, Acharya, K.R, Sturrock, E.D. | | Deposit date: | 2006-06-06 | | Release date: | 2006-10-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure of Testis Ace Glycosylation Mutants and Evidence for Conserved Domain Movement.
Biochemistry, 45, 2006
|
|
2ACY
 
 | |
2IUX
 
 | | Human tACE mutant g1234 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ANGIOTENSIN-CONVERTING ENZYME, ... | | Authors: | Watermeyer, J.M, Swell, B.T, Natesh, R, Corradi, H.R, Acharya, K.R, Sturrock, E.D. | | Deposit date: | 2006-06-07 | | Release date: | 2006-10-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Testis Ace Glycosylation Mutants and Evidence for Conserved Domain Movement.
Biochemistry, 45, 2006
|
|
2FY1
 
 | | A dual mode of RNA recognition by the RBMY protein | | Descriptor: | RNA-binding motif protein, Y chromosome, family 1 member A1, ... | | Authors: | Skrisovska, L, Bourgois, C, Stefl, R, Kister, L, Wenter, P, Elliot, D, Stevenin, J, Allain, F.H.T. | | Deposit date: | 2006-02-07 | | Release date: | 2007-02-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The testis-specific human protein RBMY recognizes RNA through a novel mode of interaction.
EMBO Rep., 8, 2007
|
|
3AFA
 
 | | The human nucleosome structure | | Descriptor: | 146-MER DNA, CHLORIDE ION, Histone H2A type 1-B/E, ... | | Authors: | Tachiwana, H, Kagawa, W, Osakabe, A, Koichiro, K, Shiga, T, Kimura, H, Kurumizaka, H. | | Deposit date: | 2010-02-24 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of instability of the nucleosome containing a testis-specific histone variant, human H3T
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2PZV
 
 | | Crystal Structure of Ketosteroid Isomerase D40N from Pseudomonas Putida (pksi) with bound Phenol | | Descriptor: | PHENOL, Steroid Delta-isomerase | | Authors: | Pybus, B, Caaveiro, J.M.M, Petsko, G.A, Ringe, D. | | Deposit date: | 2007-05-18 | | Release date: | 2007-06-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Testing Electrostatic complementarity in Enzyme Catalysis: Hydrogen Bonding in the Ketosteroid Isomerase Oxyanion Hole
PLoS Biol., 4, 2006
|
|
3DEY
 
 | |
5WBS
 
 | | Crystal structure of Frizzled-7 CRD with an inhibitor peptide Fz7-21 | | Descriptor: | Frizzled-7,inhibitor peptide Fz7-21 | | Authors: | Nile, A.H, Mukund, S, Hannoush, R.N, Wang, W. | | Deposit date: | 2017-06-29 | | Release date: | 2018-04-18 | | Last modified: | 2018-05-30 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | A selective peptide inhibitor of Frizzled 7 receptors disrupts intestinal stem cells.
Nat. Chem. Biol., 14, 2018
|
|
3DHE
 
 | | ESTROGENIC 17-BETA HYDROXYSTEROID DEHYDROGENASE COMPLEXED DEHYDROEPIANDROSTERONE | | Descriptor: | 3-BETA-HYDROXY-5-ANDROSTEN-17-ONE, ESTROGENIC 17-BETA HYDROXYSTEROID DEHYDROGENASE | | Authors: | Han, Q, Campbell, R.L, Gangloff, A, Lin, S.X. | | Deposit date: | 1998-03-25 | | Release date: | 1999-09-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Dehydroepiandrosterone and dihydrotestosterone recognition by human estrogenic 17beta-hydroxysteroid dehydrogenase. C-18/c-19 steroid discrimination and enzyme-induced strain.
J.Biol.Chem., 275, 2000
|
|
6L69
 
 | | Crystal structure of CYP154C2 from Streptomyces avermitilis | | Descriptor: | Cytochrome P450 hydroxylase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Xu, L.H, Fushinobu, S. | | Deposit date: | 2019-10-28 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Regio- and stereoselective hydroxylation of testosterone by a novel cytochrome P450 154C2 from Streptomyces avermitilis.
Biochem.Biophys.Res.Commun., 522, 2020
|
|
4L7K
 
 | | Crystal Structure of Ketosteroid Isomerase D38E from Pseudomonas Testosteroni (tKSI) | | Descriptor: | GLYCEROL, SULFATE ION, Steroid Delta-isomerase | | Authors: | Gonzalez, A, Tsai, Y, Schwans, J, Sunden, F, Herschlag, D. | | Deposit date: | 2013-06-14 | | Release date: | 2013-07-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Use of anion-aromatic interactions to position the general base in the ketosteroid isomerase active site.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3CHL
 
 | | Crystal Structure of Alpha-14 Giardin with magnesium bound | | Descriptor: | Alpha-14 giardin, MAGNESIUM ION | | Authors: | Pathuri, P, Luecke, H. | | Deposit date: | 2008-03-10 | | Release date: | 2009-01-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Apo and calcium-bound crystal structures of cytoskeletal protein alpha-14 giardin (annexin E1) from the intestinal protozoan parasite Giardia lamblia
J.Mol.Biol., 385, 2009
|
|
4BHD
 
 | |
8HPP
 
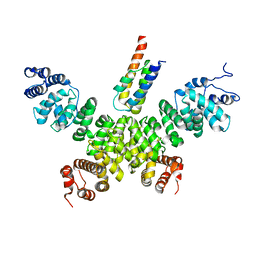 | | Crystal structure of human INTS3 with SAGE1 | | Descriptor: | Integrator complex subunit 3, Sarcoma antigen 1 | | Authors: | Deng, W, Wu, J, Lei, M. | | Deposit date: | 2022-12-12 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Cancer-testis antigen SAGE1 is a pan-cancer master regulator of RNA polymerase II
To Be Published
|
|
7FHR
 
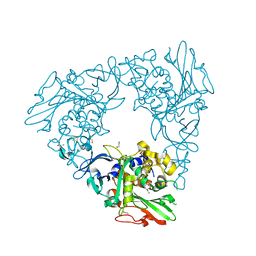 | | Crystal Structure of a Rieske Oxygenase from Cupriavidus metallidurans | | Descriptor: | 1,2-ETHANEDIOL, FE (II) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Mahto, J.K, Dhankhar, P, Kumar, P. | | Deposit date: | 2021-07-30 | | Release date: | 2021-12-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Molecular insights into substrate recognition and catalysis by phthalate dioxygenase from Comamonas testosteroni.
J.Biol.Chem., 297, 2021
|
|
