7JY3
 
 | | Structure of HbA with compound 23 (PF-07059013) | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, 6-{(1S)-1-[(2-amino-6-fluoroquinolin-3-yl)oxy]ethyl}-5-(1H-pyrazol-1-yl)pyridin-2(1H)-one, Hemoglobin subunit alpha, ... | | Authors: | Jasti, J. | | Deposit date: | 2020-08-28 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | PF-07059013: A Noncovalent Modulator of Hemoglobin for Treatment of Sickle Cell Disease.
J.Med.Chem., 64, 2021
|
|
7K0F
 
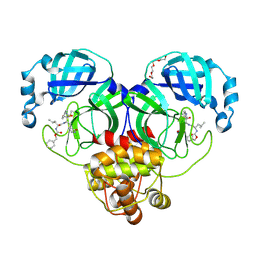 | | 1.65 A resolution structure of SARS-CoV-2 3CL protease in complex with a deuterated GC376 alpha-ketoamide analog (compound 5) | | Descriptor: | 3C-like proteinase, N-{(2S,3R)-4-(benzylamino)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-N~2~-[(benzyloxy)carbonyl]-L-leucinamide, TETRAETHYLENE GLYCOL | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Nguyen, H.N, Kim, Y, Chang, K.O, Groutas, W.C. | | Deposit date: | 2020-09-04 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Postinfection treatment with a protease inhibitor increases survival of mice with a fatal SARS-CoV-2 infection.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7K4Y
 
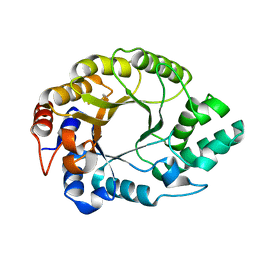 | | Crystal structure of Kemp Eliminase HG3.17 at 343 K | | Descriptor: | Endo-1,4-beta-xylanase | | Authors: | Padua, R.A.P, Otten, R, Bunzel, A, Nguyen, V, Pitsawong, W, Patterson, M, Sui, S, Perry, S.L, Cohen, A.E, Hilvert, D, Kern, D. | | Deposit date: | 2020-09-16 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | How directed evolution reshapes the energy landscape in an enzyme to boost catalysis.
Science, 370, 2020
|
|
7K7D
 
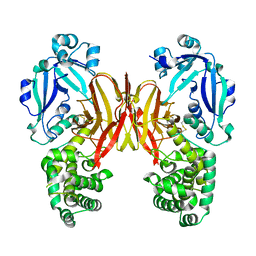 | | Crystal structure of diphtheria toxin from crystals obtained at pH 6.0 | | Descriptor: | Diphtheria toxin | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Rodnin, M.V, Ladokhin, A.S. | | Deposit date: | 2020-09-22 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the Diphtheria Toxin at Acidic pH: Implications for the Conformational Switching of the Translocation Domain.
Toxins, 12, 2020
|
|
7K82
 
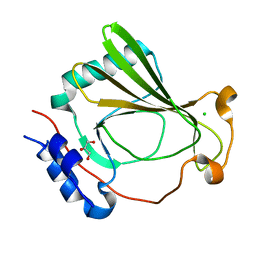 | | The X-ray crystal structure of SSR4, an S. pombe chromatin remodelling protein: sulfur SAD | | Descriptor: | CHLORIDE ION, GLYCEROL, SWI/SNF and RSC complexes subunit ssr4 | | Authors: | Peat, T.S, Newman, J. | | Deposit date: | 2020-09-24 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The X-ray crystal structure of the N-terminal domain of Ssr4, a Schizosaccharomyces pombe chromatin-remodelling protein.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
7K4X
 
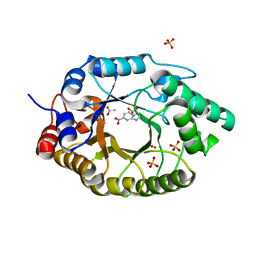 | | Crystal structure of Kemp Eliminase HG3.7 in complex with the transition state analog 6-nitrobenzotriazole | | Descriptor: | 6-NITROBENZOTRIAZOLE, ACETATE ION, Endo-1,4-beta-xylanase, ... | | Authors: | Padua, R.A.P, Otten, R, Bunzel, A, Nguyen, V, Pitsawong, W, Patterson, M, Sui, S, Perry, S.L, Cohen, A.E, Hilvert, D, Kern, D. | | Deposit date: | 2020-09-16 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | How directed evolution reshapes the energy landscape in an enzyme to boost catalysis.
Science, 370, 2020
|
|
7K66
 
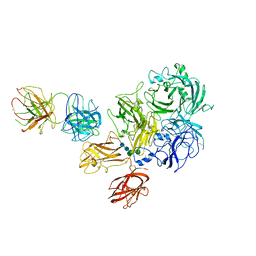 | | Structure of Blood Coagulation Factor VIII in Complex with an Anti-C1 Domain Pathogenic Antibody Inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2A9 heavy chain, ... | | Authors: | Childers, K.C, Gish, J, Jarvis, L, Peters, S, Garrels, C, Smith, I.W, Spencer, H.T, Spiegel, P.C. | | Deposit date: | 2020-09-18 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.92 Å) | | Cite: | Structure of blood coagulation factor VIII in complex with an anti-C1 domain pathogenic antibody inhibitor.
Blood, 137, 2021
|
|
7K8Q
 
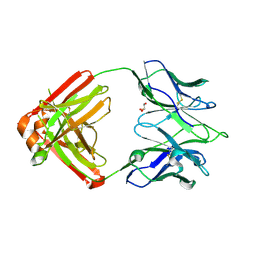 | | Crystal structure of an anti-SARS-CoV-2 human neutralizing antibody Fab fragment, C121 | | Descriptor: | C121 Fab Heavy Chain, C121 Fab Light Chain, GLYCEROL | | Authors: | Abernathy, M.E, Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2020-09-27 | | Release date: | 2020-10-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies.
Nature, 588, 2020
|
|
7JUH
 
 | |
7JVO
 
 | |
7JX7
 
 | | BRD2-BD2 in complex with a diacetylated-H2A.Z peptide | | Descriptor: | Bromodomain-containing protein 2, Diacetylated-H2A.Z peptide | | Authors: | Patel, K, Low, J.K.K, Mackay, J.P, Solomon, P.D. | | Deposit date: | 2020-08-26 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The bromodomains of BET family proteins can recognize diacetylated histone H2A.Z.
Protein Sci., 30, 2021
|
|
7K26
 
 | |
7K4Q
 
 | | Crystal structure of Kemp Eliminase HG3 in complex with the transition state analog 6-nitrobenzotriazole | | Descriptor: | 6-NITROBENZOTRIAZOLE, Endo-1,4-beta-xylanase, SULFATE ION | | Authors: | Padua, R.A.P, Otten, R, Bunzel, A, Nguyen, V, Pitsawong, W, Patterson, M, Sui, S, Perry, S.L, Cohen, A.E, Hilvert, D, Kern, D. | | Deposit date: | 2020-09-16 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | How directed evolution reshapes the energy landscape in an enzyme to boost catalysis.
Science, 370, 2020
|
|
7K4V
 
 | | Crystal structure of Kemp Eliminase HG3.17 | | Descriptor: | Endo-1,4-beta-xylanase, TETRAETHYLENE GLYCOL | | Authors: | Padua, R.A.P, Otten, R, Bunzel, A, Nguyen, V, Pitsawong, W, Patterson, M, Sui, S, Perry, S.L, Cohen, A.E, Hilvert, D, Kern, D. | | Deposit date: | 2020-09-16 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | How directed evolution reshapes the energy landscape in an enzyme to boost catalysis.
Science, 370, 2020
|
|
7K5H
 
 | | 1.90 A resolution structure of WT BfrB from Pseudomonas aeruginosa in complex with a protein-protein interaction inhibitor KM-5-66 | | Descriptor: | 4-{[3-(3-chloro-5-hydroxyphenyl)propyl]amino}-1H-isoindole-1,3(2H)-dione, Ferroxidase, POTASSIUM ION, ... | | Authors: | Lovell, S, Battaile, K.P, Soldano, A, Punchi-Hewage, A, Meraz, K, Annor-Gyamfi, J.K, Yao, H, Bunce, R.A, Rivera, M. | | Deposit date: | 2020-09-16 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Small Molecule Inhibitors of the Bacterioferritin (BfrB)-Ferredoxin (Bfd) Complex Kill Biofilm-Embedded Pseudomonas aeruginosa Cells.
Acs Infect Dis., 7, 2021
|
|
7K6I
 
 | | Carbonic Anhydrase II complexed with 4-(2-(3-phenylureido)ethylsulfonamido)benzenesulfonamide | | Descriptor: | 4-[({2-[(phenylcarbamoyl)amino]ethyl}sulfonyl)amino]benzene-1-sulfonamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Andring, J.T, Singh, S, McKenna, R. | | Deposit date: | 2020-09-20 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.406 Å) | | Cite: | Handling drug-target selectivity: A study on ureido containing Carbonic Anhydrase inhibitors.
Eur.J.Med.Chem., 212, 2021
|
|
7K4Z
 
 | | Crystal structure of Kemp Eliminase HG3.17 in complex with the transition state analog 6-nitrobenzotriazole | | Descriptor: | 6-NITROBENZOTRIAZOLE, Endo-1,4-beta-xylanase, PENTAETHYLENE GLYCOL | | Authors: | Padua, R.A.P, Otten, R, Bunzel, A, Nguyen, V, Pitsawong, W, Patterson, M, Sui, S, Perry, S.L, Cohen, A.E, Hilvert, D, Kern, D. | | Deposit date: | 2020-09-16 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | How directed evolution reshapes the energy landscape in an enzyme to boost catalysis.
Science, 370, 2020
|
|
7H8A
 
 | | Group deposition for crystallographic fragment screening of Chikungunya virus nsP3 macrodomain -- Crystal structure of Chikungunya virus nsP3 macrodomain in complex with Z3225843273 (CHIKV_MacB-x1139) | | Descriptor: | 1~{H}-pyrazol-5-ylmethanol, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Aschenbrenner, J.C, Fairhead, M, Godoy, A.S, Balcomb, B.H, Capkin, E, Chandran, A.V, Dolci, I, Golding, M, Koekemoer, L, Lithgo, R.M, Marples, P.G, Ni, X, Oliva, G, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Fearon, D, von Delft, F. | | Deposit date: | 2024-04-26 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Group deposition for crystallographic fragment screening of Chikungunya virus nsP3 macrodomain
To Be Published
|
|
7H8K
 
 | | Group deposition for crystallographic fragment screening of Chikungunya virus nsP3 macrodomain -- Crystal structure of Chikungunya virus nsP3 macrodomain in complex with Z56791928 (CHIKV_MacB-x1203) | | Descriptor: | 1-methyl-1H-benzimidazol-2-amine, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Aschenbrenner, J.C, Fairhead, M, Godoy, A.S, Balcomb, B.H, Capkin, E, Chandran, A.V, Dolci, I, Golding, M, Koekemoer, L, Lithgo, R.M, Marples, P.G, Ni, X, Oliva, G, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Fearon, D, von Delft, F. | | Deposit date: | 2024-04-26 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Group deposition for crystallographic fragment screening of Chikungunya virus nsP3 macrodomain
To Be Published
|
|
7H90
 
 | | Group deposition for crystallographic fragment screening of Chikungunya virus nsP3 macrodomain -- Crystal structure of Chikungunya virus nsP3 macrodomain in complex with Z1378432729 (CHIKV_MacB-x1337) | | Descriptor: | 1-benzofuran-2-sulfonamide, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Aschenbrenner, J.C, Fairhead, M, Godoy, A.S, Balcomb, B.H, Capkin, E, Chandran, A.V, Dolci, I, Golding, M, Koekemoer, L, Lithgo, R.M, Marples, P.G, Ni, X, Oliva, G, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Fearon, D, von Delft, F. | | Deposit date: | 2024-04-26 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Group deposition for crystallographic fragment screening of Chikungunya virus nsP3 macrodomain
To Be Published
|
|
7H9C
 
 | | Group deposition for crystallographic fragment screening of Chikungunya virus nsP3 macrodomain -- Crystal structure of Chikungunya virus nsP3 macrodomain in complex with Z31721188 (CHIKV_MacB-x1421) | | Descriptor: | (3-hydroxyphenyl)(morpholin-4-yl)methanone, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Aschenbrenner, J.C, Fairhead, M, Godoy, A.S, Balcomb, B.H, Capkin, E, Chandran, A.V, Dolci, I, Golding, M, Koekemoer, L, Lithgo, R.M, Marples, P.G, Ni, X, Oliva, G, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Fearon, D, von Delft, F. | | Deposit date: | 2024-04-26 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Group deposition for crystallographic fragment screening of Chikungunya virus nsP3 macrodomain
To Be Published
|
|
7JO3
 
 | | Carbonic Anhydrase IX Mimic Complexed with N-(5-sulfamoyl-1,3,4-thiadiazol-2-yl)pivalamide | | Descriptor: | 2,2-dimethyl-N-(5-sulfamoyl-1,3,4-thiadiazol-2-yl)propanamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Andring, J.T, McKenna, R. | | Deposit date: | 2020-08-05 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.454 Å) | | Cite: | Structural Basis of Nanomolar Inhibition of Tumor-Associated Carbonic Anhydrase IX: X-Ray Crystallographic and Inhibition Study of Lipophilic Inhibitors with Acetazolamide Backbone.
J.Med.Chem., 63, 2020
|
|
7JTW
 
 | | Crystal structure of RORgt with compound (4R)-6-[(2,5-dichloro-3-{[(2R,4R)-1-(cyclopentanecarbonyl)-2-methylpiperidin-4-yl]oxy}phenyl)amino]-6-oxo-4-phenylhexanoic acid | | Descriptor: | (4R)-6-[(2,5-dichloro-3-{[(2R,4R)-1-(cyclopentanecarbonyl)-2-methylpiperidin-4-yl]oxy}phenyl)amino]-6-oxo-4-phenylhexanoic acid, GLYCEROL, RAR-related orphan receptor C isoform a variant, ... | | Authors: | Min, X, Wang, Z. | | Deposit date: | 2020-08-18 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of 6-Oxo-4-phenyl-hexanoic acid derivatives as ROR gamma t inverse agonists showing favorable ADME profile.
Bioorg.Med.Chem.Lett., 36, 2021
|
|
7K7E
 
 | | Crystal structure of diphtheria toxin from crystals obtained at pH 7.0 | | Descriptor: | Diphtheria toxin | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Rodnin, M.V, Ladokhin, A.S. | | Deposit date: | 2020-09-22 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Diphtheria Toxin at Acidic pH: Implications for the Conformational Switching of the Translocation Domain.
Toxins, 12, 2020
|
|
7JNO
 
 | |
