4D8B
 
 | |
5WCX
 
 | |
5BP8
 
 | | ent-Copalyl diphosphate synthase from Streptomyces platensis | | Descriptor: | 1,2-ETHANEDIOL, Ent-copalyl diphosphate synthase, SULFATE ION | | Authors: | Osipiuk, J, Hatzos-Skintges, C, Endres, M, Babnigg, G, Rudolf, J.D, Ma, M, Chang, C.-Y, Shen, B, Joachimiak, A, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-05-27 | | Release date: | 2015-06-10 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the ent-Copalyl Diphosphate Synthase PtmT2 from Streptomyces platensis CB00739, a Bacterial Type II Diterpene Synthase.
J.Am.Chem.Soc., 138, 2016
|
|
6UZ5
 
 | |
3ZH4
 
 | |
5A0L
 
 | | N-terminal thioester domain of fibronectin-binding protein SfbI from Streptococcus pyogenes | | Descriptor: | ACETATE ION, FIBRONECTIN-BINDING PROTEIN, ZINC ION | | Authors: | Walden, M, Edwards, J.M, Dziewulska, A.M, Kan, S.-Y, Schwarz-Linek, U, Banfield, M.J. | | Deposit date: | 2015-04-21 | | Release date: | 2015-06-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | An internal thioester in a pathogen surface protein mediates covalent host binding.
Elife, 4, 2015
|
|
3QSP
 
 | | Analysis of a new family of widely distributed metal-independent alpha mannosidases provides unique insight into the processing of N-linked glycans, Streptococcus pneumoniae SP_2144 non-productive substrate complex with alpha-1,6-mannobiose | | Descriptor: | 1,2-ETHANEDIOL, Putative uncharacterized protein, alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose | | Authors: | Gregg, K.J, Zandberg, W.F, Hehemann, J.-H, Whitworth, G.E, Deng, L.E, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2011-02-21 | | Release date: | 2011-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Analysis of a New Family of Widely Distributed Metal-independent {alpha}-Mannosidases Provides Unique Insight into the Processing of N-Linked Glycans.
J.Biol.Chem., 286, 2011
|
|
6VJB
 
 | |
3ZPP
 
 | |
5XYA
 
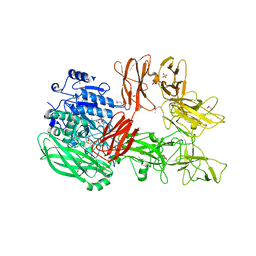 | | Crystal structure of a serine protease from Streptococcus species | | Descriptor: | 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, CALCIUM ION, Chemokine protease C, ... | | Authors: | Jobichen, C, Sivaraman, J. | | Deposit date: | 2017-07-06 | | Release date: | 2018-08-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of ScpC, a virulence protease fromStreptococcus pyogenes, reveals the functional domains and maturation mechanism.
Biochem. J., 475, 2018
|
|
6VPC
 
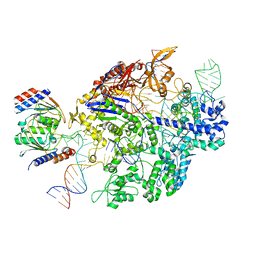 | | Structure of the SpCas9 DNA adenine base editor - ABE8e | | Descriptor: | CRISPR-associated endonuclease Cas9, Cas9 (SpCas9) single-guide RNA (sgRNA), DNA non-target strand (NTS), ... | | Authors: | Knott, G.J, Lapinaite, A, Doudna, J.A. | | Deposit date: | 2020-02-03 | | Release date: | 2020-07-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | DNA capture by a CRISPR-Cas9-guided adenine base editor.
Science, 369, 2020
|
|
5AOU
 
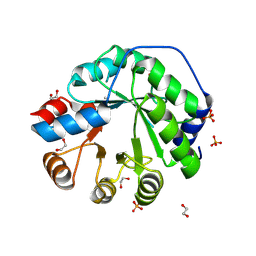 | | Structure of the engineered retro-aldolase RA95.5-8F apo | | Descriptor: | 1,2-ETHANEDIOL, INDOLE-3-GLYCEROL PHOSPHATE SYNTHASE, PHOSPHATE ION | | Authors: | Obexer, R, Mittl, P, Hilvert, D. | | Deposit date: | 2015-09-11 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Emergence of a catalytic tetrad during evolution of a highly active artificial aldolase.
Nat Chem, 9, 2017
|
|
3GI1
 
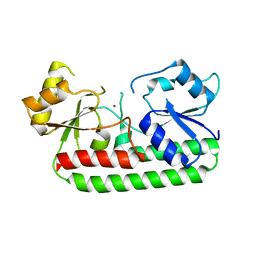 | | Crystal Structure of the laminin-binding protein Lbp of Streptococcus pyogenes | | Descriptor: | Laminin-binding protein of group A streptococci, ZINC ION | | Authors: | Linke, C, Caradoc-Davies, T.T, Young, P.G, Proft, T, Baker, E.N. | | Deposit date: | 2009-03-04 | | Release date: | 2009-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The laminin-binding protein Lbp from Streptococcus pyogenes is a zinc receptor
J.Bacteriol., 191, 2009
|
|
4CU7
 
 | | Unravelling the multiple functions of the architecturally intricate Streptococcus pneumoniae beta-galactosidase, BgaA | | Descriptor: | 1,2-ETHANEDIOL, BETA-GALACTOSIDASE, D-galacto-isofagomine, ... | | Authors: | Singh, A.K, Pluvinage, B, Higgins, M.A, Dalia, A.B, Flynn, M, Lloyd, A.R, Weiser, J.N, Stubbs, K.A, Boraston, A.B, King, S.J. | | Deposit date: | 2014-03-17 | | Release date: | 2014-08-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Unravelling the Multiple Functions of the Architecturally Intricate Streptococcus Pneumoniae Beta-Galactosidase, BgaA.
Plos Pathog., 10, 2014
|
|
4CGK
 
 | | Crystal structure of the essential protein PcsB from Streptococcus pneumoniae | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Bartual, S.G, Straume, D, Stamsas, G.A, Alfonso, C, Martinez-Ripoll, M, Havarstein, L.S, Hermoso, J.A. | | Deposit date: | 2013-11-25 | | Release date: | 2014-05-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Basis of Pcsb-Mediated Cell Separation in Streptococcus Pneumoniae.
Nat.Commun., 5, 2014
|
|
3PN9
 
 | |
3PYL
 
 | | Crystal structure of aspartate beta-semialdehide dehydrogenase from Streptococcus pneumoniae with D-2,3-diaminopropionate | | Descriptor: | 3-amino-D-alanine, Aspartate-semialdehyde dehydrogenase | | Authors: | Pavlovsky, A.G, Liu, X, Faehnle, C.R, Potente, N, Viola, R.E. | | Deposit date: | 2010-12-13 | | Release date: | 2012-01-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Characterization of Inhibitors with Selectivity against Members of a Homologous Enzyme Family.
Chem.Biol.Drug Des., 79, 2012
|
|
5CR6
 
 | | Structure of pneumolysin at 1.98 A resolution | | Descriptor: | Pneumolysin | | Authors: | Marshall, J.E, Faraj, B.H.A, Gingras, A.R, Lonnen, R, Sheikh, M.A, El-Mezgueldi, M, Moody, P.C.E, Andrew, P.W, Wallis, R. | | Deposit date: | 2015-07-22 | | Release date: | 2015-09-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The Crystal Structure of Pneumolysin at 2.0 angstrom Resolution Reveals the Molecular Packing of the Pre-pore Complex.
Sci Rep, 5, 2015
|
|
3PZB
 
 | | Crystals Structure of Aspartate beta-Semialdehyde Dehydrogenase complex with NADP and D-2,3-Diaminopropionate | | Descriptor: | 1,2-ETHANEDIOL, 3-amino-D-alanine, Aspartate-semialdehyde dehydrogenase, ... | | Authors: | Pavlovsky, A.G, Viola, R.E. | | Deposit date: | 2010-12-14 | | Release date: | 2012-01-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Characterization of Inhibitors with Selectivity against Members of a Homologous Enzyme Family.
Chem.Biol.Drug Des., 79, 2012
|
|
4BUG
 
 | | Pilus-presented adhesin, Spy0125 (Cpa), Cys426Ala mutant | | Descriptor: | ANCILLARY PROTEIN 1 | | Authors: | Walden, M, Crow, A, Nelson, M, Banfield, M.J. | | Deposit date: | 2013-06-20 | | Release date: | 2013-10-09 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Intramolecular Isopeptide But not Internal Thioester Bonds Confer Proteolytic and Significant Thermal Stability to the S. Pyogenes Pilus Adhesin Spy0125.
Proteins, 82, 2014
|
|
5A35
 
 | | Crystal structure of Glycine Cleavage Protein H-Like (GcvH-L) from Streptococcus pyogenes | | Descriptor: | GLYCINE CLEAVAGE SYSTEM H PROTEIN, PENTAETHYLENE GLYCOL | | Authors: | Rack, J.G.M, Morra, R, Barkauskaite, E, Kraehenbuehl, R, Ariza, A, Qu, Y, Ortmayer, M, Leidecker, O, Cameron, D.R, Matic, I, Peleg, A.Y, Leys, D, Traven, A, Ahel, I. | | Deposit date: | 2015-05-27 | | Release date: | 2015-07-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Identification of a Class of Protein Adp-Ribosylating Sirtuins in Microbial Pathogens.
Mol.Cell, 59, 2015
|
|
5A3C
 
 | | Crystal structure of the ADP-ribosylating sirtuin (SirTM) from Streptococcus pyogenes in complex with NAD | | Descriptor: | 1,2-ETHANEDIOL, GLYCINE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Rack, J.G.M, Morra, R, Barkauskaite, E, Kraehenbuehl, R, Ariza, A, Qu, Y, Ortmayer, M, Leidecker, O, Cameron, D.R, Matic, I, Peleg, A.Y, Leys, D, Traven, A, Ahel, I. | | Deposit date: | 2015-05-28 | | Release date: | 2015-07-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Identification of a Class of Protein Adp-Ribosylating Sirtuins in Microbial Pathogens.
Mol.Cell, 59, 2015
|
|
4C0Z
 
 | | The N-terminal domain of the Streptococcus pyogenes pilus tip adhesin Cpa | | Descriptor: | ANCILLARY PROTEIN 2, CHLORIDE ION, GLYCEROL, ... | | Authors: | Linke-Winnebeck, C, Paterson, N, Baker, E.N. | | Deposit date: | 2013-08-08 | | Release date: | 2013-11-20 | | Last modified: | 2014-01-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Model for the Covalent Adhesion of the Streptococcus Pyogenes Pilus Through a Thioester Bond.
J.Biol.Chem., 289, 2014
|
|
4D0Y
 
 | |
4CP6
 
 | | The Crystal structure of Pneumococcal vaccine antigen PcpA | | Descriptor: | CHOLINE BINDING PROTEIN PCPA | | Authors: | Vallee, F, Steier, V, Oloo, E, Chawla, D, Vonrhein, C, Steinmetz, A, Mathieu, M, Rak, A, Mikol, V, Oomen, R. | | Deposit date: | 2014-01-31 | | Release date: | 2015-02-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | The Crystal Structure of Pneumoccocal Vaccine Antigen Pcpa
To be Published
|
|
