2RR9
 
 | | The solution structure of the K63-Ub2:tUIMs complex | | Descriptor: | Putative uncharacterized protein UIMC1, ubiquitin | | Authors: | Sekiyama, N, Jee, J, Isogai, S, Akagi, K, Huang, T, Ariyoshi, M, Tochio, H, Shirakawa, M. | | Deposit date: | 2010-06-16 | | Release date: | 2011-07-06 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the K63-Ub2:tUIMs complex
To be Published
|
|
4B1X
 
 | | Structure of the Phactr1 RPEL-2 bound to G-actin | | Descriptor: | ACTIN, ALPHA SKELETAL MUSCLE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Mouilleron, S, Wiezlak, M, O'Reilly, N, Treisman, R, McDonald, N.Q. | | Deposit date: | 2012-07-12 | | Release date: | 2013-07-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of the Phactr1 RPEL domain and RPEL motif complexes with G-actin reveal the molecular basis for actin binding cooperativity.
Structure, 20, 2012
|
|
7ZET
 
 | | Crystal structure of human Clusterin, crystal form I | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Clusterin, ... | | Authors: | Yuste-Checa, P, Bracher, A, Hartl, F.U. | | Deposit date: | 2022-03-31 | | Release date: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of human Clusterin, crystal form I
To be published
|
|
7ZEU
 
 | |
1EXA
 
 | | ENANTIOMER DISCRIMINATION ILLUSTRATED BY CRYSTAL STRUCTURES OF THE HUMAN RETINOIC ACID RECEPTOR HRARGAMMA LIGAND BINDING DOMAIN: THE COMPLEX WITH THE ACTIVE R-ENANTIOMER BMS270394. | | Descriptor: | DODECYL-ALPHA-D-MALTOSIDE, R-3-FLUORO-4-[2-HYDROXY-2-(5,5,8,8-TETRAMETHYL-5,6,7,8,-TETRAHYDRO-NAPHTALEN-2-YL)-ACETYLAMINO]-BENZOIC ACID, RETINOIC ACID RECEPTOR GAMMA-2 | | Authors: | Klaholz, B.P, Mitschler, A, Belema, M, Zusi, C, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2000-05-02 | | Release date: | 2000-06-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Enantiomer discrimination illustrated by high-resolution crystal structures of the human nuclear receptor hRARgamma.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
7YO2
 
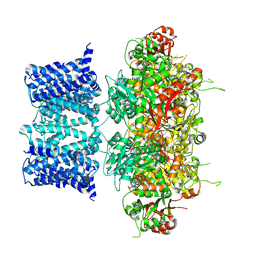 | |
4B1Y
 
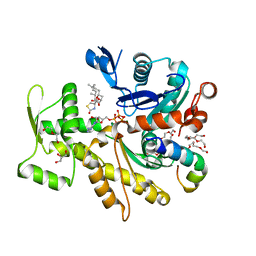 | | Structure of the Phactr1 RPEL-3 bound to G-actin | | Descriptor: | ACTIN, ALPHA SKELETAL MUSCLE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Mouilleron, S, Wiezlak, M, O'Reilly, N, Treisman, R, McDonald, N.Q. | | Deposit date: | 2012-07-12 | | Release date: | 2013-07-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Structures of the Phactr1 RPEL domain and RPEL motif complexes with G-actin reveal the molecular basis for actin binding cooperativity.
Structure, 20, 2012
|
|
7YO4
 
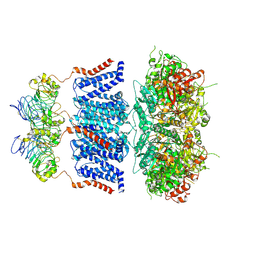 | |
7YO5
 
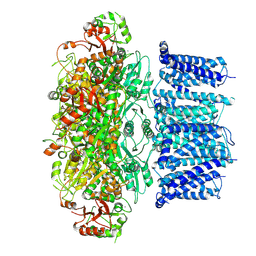 | |
7YNZ
 
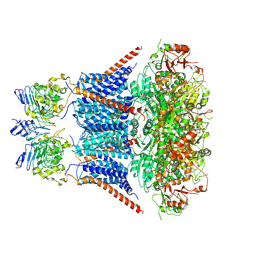 | |
7YO1
 
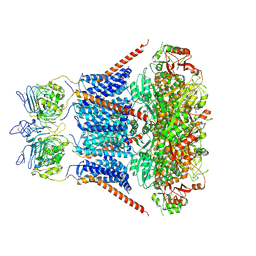 | | Cryo-EM structure of RCK1 mutated human Slo1-LRRC26 complex | | Descriptor: | CALCIUM ION, Calcium-activated potassium channel subunit alpha-1, Leucine-rich repeat-containing protein 26, ... | | Authors: | Yamanouchi, D, Nureki, O. | | Deposit date: | 2022-08-01 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for dual allosteric gating modulation of Slo1-LRRC channel complex
To Be Published
|
|
7YO0
 
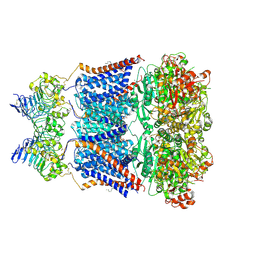 | | Cryo-EM structure of human Slo1-LRRC26 complex with Symmetry Expansion | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, CALCIUM ION, Calcium-activated potassium channel subunit alpha-1, ... | | Authors: | Yamanouchi, D, Nureki, O. | | Deposit date: | 2022-08-01 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for dual allosteric gating modulation of Slo1-LRRC channel complex
To Be Published
|
|
6A3A
 
 | | MVM NES mutant Nm2 in complex with CRM1-Ran-RanBP1 | | Descriptor: | 1,2-ETHANEDIOL, Exportin-1, GLYCEROL, ... | | Authors: | Sun, Q, Li, Y. | | Deposit date: | 2018-06-15 | | Release date: | 2019-06-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cancer Therapy with Nanoparticle-Medicated Intracellular Expression of Peptide CRM1-Inhibitor.
Int J Nanomedicine, 16, 2021
|
|
2RQS
 
 | | 3D structure of Pin from the psychrophilic archeon Cenarcheaum symbiosum (CsPin) | | Descriptor: | Parvulin-like peptidyl-prolyl isomerase | | Authors: | Zhukov, I, Jaremko, L, Jaremko, M, Mueller, J.W, Bayer, P. | | Deposit date: | 2009-11-17 | | Release date: | 2010-11-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and Dynamics of the First Archaeal Parvulin Reveal a New Functionally Important Loop in Parvulin-type Prolyl Isomerases
J.Biol.Chem., 286, 2011
|
|
6A3E
 
 | | MVM NES mutant Nm15 in complex with CRM1-Ran-RanBP1 | | Descriptor: | Exportin-1, GTP-binding nuclear protein Ran, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Sun, Q, Li, Y. | | Deposit date: | 2018-06-15 | | Release date: | 2019-06-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Cancer Therapy with Nanoparticle-Medicated Intracellular Expression of Peptide CRM1-Inhibitor.
Int J Nanomedicine, 16, 2021
|
|
3NKH
 
 | | Crystal Structure of Integrase from MRSA strain Staphylococcus aureus | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, ... | | Authors: | Kim, Y, Tesar, C, Gornicki, P, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-06-19 | | Release date: | 2010-08-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Crystal Structure of Integrase from MRSA strain Staphylococcus aureus
To be Published, 2010
|
|
1EXX
 
 | | ENANTIOMER DISCRIMINATION ILLUSTRATED BY CRYSTAL STRUCTURES OF THE HUMAN RETINOIC ACID RECEPTOR HRARGAMMA LIGAND BINDING DOMAIN: THE COMPLEX WITH THE INACTIVE S-ENANTIOMER BMS270395. | | Descriptor: | 3-FLUORO-4-[2-HYDROXY-2-(5,5,8,8-TETRAMETHYL-5,6,7,8,-TETRAHYDRO-NAPHTALEN-2-YL)-ACETYLAMINO]-BENZOIC ACID, DODECYL-ALPHA-D-MALTOSIDE, RETINOIC ACID RECEPTOR GAMMA-2 | | Authors: | Klaholz, B.P, Mitschler, A, Belema, M, Zusi, C, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2000-05-05 | | Release date: | 2000-06-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Enantiomer discrimination illustrated by high-resolution crystal structures of the human nuclear receptor hRARgamma.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
5XSO
 
 | | Crystal structure of full-length FixJ from B. japonicum crystallized in space group C2221 | | Descriptor: | FORMIC ACID, GLYCEROL, Response regulator FixJ | | Authors: | Nishizono, Y, Hisano, T, Sawai, H, Shiro, Y, Nakamura, H, Wright, G.S.A, Saeki, A, Hikima, T, Yamamoto, M, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2017-06-14 | | Release date: | 2018-05-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.778 Å) | | Cite: | Architecture of the complete oxygen-sensing FixL-FixJ two-component signal transduction system.
Sci Signal, 11, 2018
|
|
4POJ
 
 | | Crystal structure of human Retinoid X Receptor alpha-ligand binding domain complex with 7-methyl UAB30 and the coactivator peptide GRIP-1 | | Descriptor: | (2E,4E,6Z,8E)-3,7-dimethyl-8-(7-methyl-3,4-dihydronaphthalen-1(2H)-ylidene)octa-2,4,6-trienoic acid, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | Xia, G, Smith, C.D, Muccio, D.D. | | Deposit date: | 2014-02-25 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Methyl substitution of a rexinoid agonist improves potency and reveals site of lipid toxicity.
J.Med.Chem., 57, 2014
|
|
7ZH9
 
 | | Uba1 in complex with ATP | | Descriptor: | ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Misra, M, Schindelin, H. | | Deposit date: | 2022-04-05 | | Release date: | 2022-08-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structures of UBA6 explain its dual specificity for ubiquitin and FAT10.
Nat Commun, 13, 2022
|
|
6A7P
 
 | | Human serum albumin complexed with aripiprazole | | Descriptor: | 7-[4-[4-[2,3-bis(chloranyl)phenyl]piperazin-1-yl]butoxy]-3,4-dihydro-1H-quinolin-2-one, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Kawai, A, Yamasaki, K, Otagiri, M. | | Deposit date: | 2018-07-03 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Analysis of the Binding of Aripiprazole to Human Serum Albumin: The Importance of a Chloro-Group in the Chemical Structure.
Acs Omega, 3, 2018
|
|
2ST1
 
 | |
2RSD
 
 | | Solution structure of the plant homeodomain (PHD) of the E3 SUMO ligase Siz1 from rice | | Descriptor: | E3 SUMO-protein ligase SIZ1, ZINC ION | | Authors: | Shindo, H, Tsuchiya, W, Suzuki, R, Yamazaki, T. | | Deposit date: | 2012-01-12 | | Release date: | 2012-08-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | PHD finger of the SUMO ligase Siz/PIAS family in rice reveals specific binding for methylated histone H3 at lysine 4 and arginine 2
Febs Lett., 586, 2012
|
|
7EWJ
 
 | | Toxin-antitoxin complex from Staphylococcus aureus | | Descriptor: | Endoribonuclease MazF, GLYCEROL, PemI inhibitor, ... | | Authors: | Kim, D.H, Kang, S.M, Lee, S.J, Lee, B.J. | | Deposit date: | 2021-05-25 | | Release date: | 2022-02-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Role of PemI in the Staphylococcus aureus PemIK toxin-antitoxin complex: PemI controls PemK by acting as a PemK loop mimic.
Nucleic Acids Res., 50, 2022
|
|
8A8C
 
 | | T5 phage receptor-binding protein pb5 bound to ferrichrome transporter FhuA | | Descriptor: | Ferrichrome outer membrane transporter/phage receptor, Receptor-binding protein pb5, [(2R,3S,4R,5R,6R)-2-[[(2R,4R,5R,6R)-6-[(1R)-1,2-bis(oxidanyl)ethyl]-4-[(2R,4R,5R,6R)-6-[(1R)-1,2-bis(oxidanyl)ethyl]-2-carboxy-4,5-bis(oxidanyl)oxan-2-yl]oxy-2-carboxy-5-oxidanyl-oxan-2-yl]oxymethyl]-5-[[(3R)-3-dodecanoyloxytetradecanoyl]amino]-4-(3-nonanoyloxypropanoyloxy)-6-[[(2R,3S,4R,5R,6R)-3-oxidanyl-4-[(3S)-3-oxidanyltetradecanoyl]oxy-5-[[(3R)-3-oxidanyltridecanoyl]amino]-6-phosphonatooxy-oxan-2-yl]methoxy]oxan-3-yl] phosphate | | Authors: | Silale, A, van den Berg, B. | | Deposit date: | 2022-06-22 | | Release date: | 2022-10-05 | | Last modified: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for host recognition and superinfection exclusion by bacteriophage T5.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
