2A9I
 
 | |
4ZOC
 
 | | Crystal Structure of mutant (D270A) beta-glucosidase from Listeria innocua in complex with sophorotriose | | Descriptor: | GLYCEROL, Lin1840 protein, MAGNESIUM ION, ... | | Authors: | Nakajima, M, Yoshida, R, Miyanaga, A, Abe, K, Takahashi, Y, Sugimoto, N, Toyoizumi, H, Nakai, H, Kitaoka, M, Taguchi, H. | | Deposit date: | 2015-05-06 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Functional and Structural Analysis of a beta-Glucosidase Involved in beta-1,2-Glucan Metabolism in Listeria innocua
Plos One, 11, 2016
|
|
3A12
 
 | | Crystal structure of Type III Rubisco complexed with 2-CABP | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, Ribulose bisphosphate carboxylase | | Authors: | Nishitani, Y, Fujihashi, M, Doi, T, Yoshida, S, Atomi, H, Imanaka, T, Miki, K. | | Deposit date: | 2009-03-25 | | Release date: | 2010-04-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-based catalytic optimization of a type III Rubisco from a hyperthermophile
J.Biol.Chem., 285, 2010
|
|
4CG4
 
 | | Crystal structure of the CHS-B30.2 domains of TRIM20 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-ETHANETHIOL, PYRIN, ... | | Authors: | Weinert, C, Morger, D, Djekic, A, Mittl, P.R.E, Gruetter, M.G. | | Deposit date: | 2013-11-20 | | Release date: | 2015-04-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Trim20 C-Terminal Coiled-Coil/B30.2 Fragment: Implications for the Recognition of Higher Order Oligomers
Sci.Rep., 5, 2015
|
|
9IIP
 
 | | Fucokinase part of FKP with beta-L-fucose 1-phosphate | | Descriptor: | 6-deoxy-1-O-phosphono-beta-L-galactopyranose, L-fucokinase/L-fucose-1-P guanylyltransferase | | Authors: | Ko, T.P, Lin, S.W, Hsu, M.F, Lin, C.H. | | Deposit date: | 2024-06-21 | | Release date: | 2025-03-05 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural insight into the catalytic mechanism of the bifunctional enzyme l-fucokinase/GDP-fucose pyrophosphorylase.
J.Biol.Chem., 301, 2025
|
|
6ID0
 
 | | Cryo-EM structure of a human intron lariat spliceosome prior to Prp43 loaded (ILS1 complex) at 2.9 angstrom resolution | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, CWF19-like protein 2, Cell division cycle 5-like protein, ... | | Authors: | Zhang, X, Zhan, X, Yan, C, Shi, Y. | | Deposit date: | 2018-09-07 | | Release date: | 2019-03-13 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of the human spliceosomes before and after release of the ligated exon.
Cell Res., 29, 2019
|
|
1Q9E
 
 | | RNase T1 variant with adenine specificity | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Guanyl-specific ribonuclease T1 precursor | | Authors: | Czaja, R, Struhalla, M, Hoeschler, K, Saenger, W, Straeter, N, Hahn, U. | | Deposit date: | 2003-08-25 | | Release date: | 2004-03-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | RNase T1 Variant RV Cleaves Single-Stranded RNA after Purines Due to Specific Recognition by the Asn46 Side Chain Amide.
Biochemistry, 43, 2004
|
|
7V7J
 
 | | Cryo-EM structure of SARS-CoV-2 S-Kappa variant (B.1.617.1), dimer of S trimer conformation 3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-08-21 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 S-Kappa variant (B.1.617.1), dimer of S trimer conformation 3
To Be Published
|
|
5EBT
 
 | | Tankyrase 1 with Phthalazinone 2 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 4-[bis(fluoranyl)-[3-[[(6~{S})-6-methyl-3-(trifluoromethyl)-6,8-dihydro-5~{H}-[1,2,4]triazolo[4,3-a]pyrazin-7-yl]carbonyl]phenyl]methyl]-2~{H}-phthalazin-1-one, Tankyrase-1, ... | | Authors: | Kazmirski, S.L, Johannes, J. | | Deposit date: | 2015-10-19 | | Release date: | 2016-03-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Discovery of AZ0108, an orally bioavailable phthalazinone PARP inhibitor that blocks centrosome clustering.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
1MWW
 
 | | THE STRUCTURE OF THE HYPOTHETICAL PROTEIN HI1388.1 FROM HAEMOPHILUS INFLUENZAE REVEALS A TAUTOMERASE/MIF FOLD | | Descriptor: | CHLORIDE ION, GLUTAMIC ACID, HYPOTHETICAL PROTEIN HI1388.1 | | Authors: | Lehmann, C, Pullalarevu, S, Krajewski, W, Galkin, A, Howard, A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2002-10-01 | | Release date: | 2003-11-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structure of the Hypothetical Protein HI1388.1 from Haemophilus influenzae
To be Published
|
|
7V78
 
 | | Cryo-EM structure of SARS-CoV-2 S-Gamma variant (P.1), one RBD-up conformation 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-08-21 | | Release date: | 2021-10-06 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 S-Gamma variant (P.1), one RBD-up conformation 1
To Be Published
|
|
7V7H
 
 | | Cryo-EM structure of SARS-CoV-2 S-Kappa variant (B.1.617.1), dimer of S trimer conformation 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-08-21 | | Release date: | 2021-10-06 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 S-Kappa variant (B.1.617.1), dimer of S trimer conformation 1
To Be Published
|
|
5DL9
 
 | | Structure of Tetragonal Lysozyme in complex with Iodine solved by UWO Students | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, IODIDE ION, ... | | Authors: | Bednarski, R, Cirricione, N, Greco, A, Hodgson, R, Kent, S, McGowan, J, Notherm, B, Patt, M, Vue, L, Bianchetti, C.M. | | Deposit date: | 2015-09-04 | | Release date: | 2015-09-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structure of Tetragonal Lysozyme in complex with Iodine solved by UWO Students
To Be Published
|
|
7BE5
 
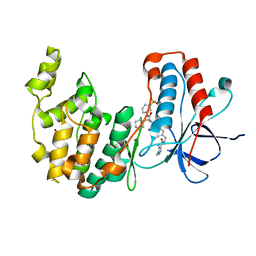 | | Crystal structure of MAP kinase p38 alpha in complex with inhibitor SR276 | | Descriptor: | 1,2-ETHANEDIOL, 5-azanyl-~{N}-[[4-[[(2~{S})-4-cyclohexyl-1-[[(3~{R})-1-methylsulfonylpiperidin-3-yl]amino]-1-oxidanylidene-butan-2-yl]carbamoyl]phenyl]methyl]-1-methyl-pyrazole-4-carboxamide, Mitogen-activated protein kinase 14 | | Authors: | Joerger, A.C, Schroeder, M, Roehm, S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-22 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8000524 Å) | | Cite: | Development of a Selective Dual Discoidin Domain Receptor (DDR)/p38 Kinase Chemical Probe.
J.Med.Chem., 64, 2021
|
|
7V7I
 
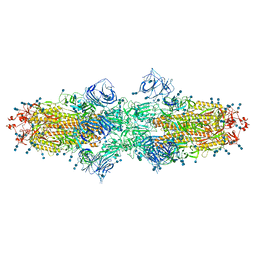 | | Cryo-EM structure of SARS-CoV-2 S-Kappa variant (B.1.617.1), dimer of S trimer conformation 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-08-21 | | Release date: | 2021-10-06 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 S-Kappa variant (B.1.617.1), dimer of S trimer conformation 2
To Be Published
|
|
7V87
 
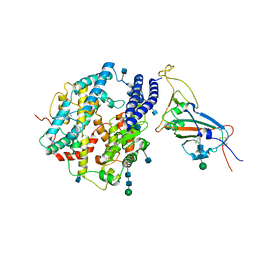 | | Local refinement of SARS-CoV-2 S-Kappa variant (B.1.617.1) RBD and Angiotensin-converting enzyme 2 (ACE2) ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2,Angiotensin-converting enzyme 2 (ACE2) ectodomain, ... | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-08-22 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Local refinement of SARS-CoV-2 S-Kappa variant (B.1.617.1) RBD and Angiotensin-converting enzyme 2 (ACE2) ectodomain
To Be Published
|
|
2VPF
 
 | |
6KK2
 
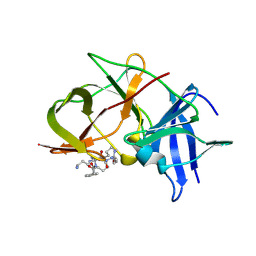 | | Crystal structure of Zika NS2B-NS3 protease with compound 2 | | Descriptor: | 1-[(10~{S},17~{S},20~{S})-17,20-bis(4-azanylbutyl)-4,9,16,19,22-pentakis(oxidanylidene)-3,8,15,18,21-pentazabicyclo[22.2.2]octacosa-1(27),24(28),25-trien-10-yl]guanidine, NS3 protease, Serine protease subunit NS2B | | Authors: | Quek, J.P. | | Deposit date: | 2019-07-23 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structure-Based Macrocyclization of Substrate Analogue NS2B-NS3 Protease Inhibitors of Zika, West Nile and Dengue viruses.
Chemmedchem, 15, 2020
|
|
5DR3
 
 | | Endothiapepsin in complex with fragment 333 | | Descriptor: | 1,2-ETHANEDIOL, 4-propan-2-ylsulfanyl-1-propyl-6,7-dihydro-5~{H}-cyclopenta[d]pyrimidin-2-one, ACETATE ION, ... | | Authors: | Schiebel, J, Heine, A, Klebe, G. | | Deposit date: | 2015-09-15 | | Release date: | 2016-09-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
2PFO
 
 | | DNA Polymerase lambda in complex with DNA and dUPNPP | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, DNA polymerase lambda, ... | | Authors: | Garcia-Diaz, M, Bebenek, K, Krahn, J.M, Pedersen, L.C, Kunkel, T.A. | | Deposit date: | 2007-04-05 | | Release date: | 2007-05-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Role of the catalytic metal during polymerization by DNA polymerase lambda.
DNA Repair, 6, 2007
|
|
8XNT
 
 | | Respiratory complex Peripheral Arm of CI, close form C, focus-refined map of type IB, Wild type mouse under Cold Acclimation | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Shin, Y.-C, Liao, M. | | Deposit date: | 2023-12-30 | | Release date: | 2024-10-30 | | Last modified: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of respiratory complex adaptation to cold temperatures.
Cell, 187, 2024
|
|
8XNY
 
 | | Respiratory complex Peripheral Arm of CI, open form B, focus-refined map of type II, Wild type mouse under Cold Acclimation | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Shin, Y.-C, Liao, M. | | Deposit date: | 2023-12-30 | | Release date: | 2024-10-30 | | Last modified: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of respiratory complex adaptation to cold temperatures.
Cell, 187, 2024
|
|
5DRY
 
 | | Crystal structure of Dot1L in complex with inhibitor CPD3 [N-(1-(2-chlorophenyl)-1H-indol-6-yl)-2-(2-(5-(2-chlorophenyl)-1H-tetrazol-1-yl)acetyl)hydrazinecarboxamide] | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-79 specific, N-[1-(2-chlorophenyl)-1H-indol-6-yl]-2-{[5-(2-chlorophenyl)-1H-tetrazol-1-yl]acetyl}hydrazinecarboxamide, ... | | Authors: | Scheufler, C, Gaul, C, Be, C, Moebitz, H. | | Deposit date: | 2015-09-16 | | Release date: | 2016-06-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Discovery of Novel Dot1L Inhibitors through a Structure-Based Fragmentation Approach.
Acs Med.Chem.Lett., 7, 2016
|
|
5U8A
 
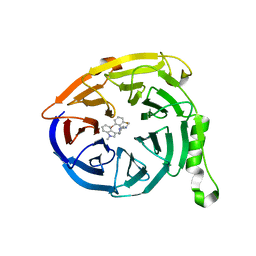 | | Polycomb protein EED in complex with inhibitor: (3R,4S)-1-[(2-bromo-6-fluorophenyl)methyl]-N,N-dimethyl-4-(1-methyl-1H-indol-3-yl)pyrrolidin-3-amine | | Descriptor: | (3R,4S)-1-[(2-bromo-6-fluorophenyl)methyl]-N,N-dimethyl-4-(1-methyl-1H-indol-3-yl)pyrrolidin-3-amine, Polycomb protein EED | | Authors: | Jakob, C.G, Zhu, H. | | Deposit date: | 2016-12-14 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | SAR of amino pyrrolidines as potent and novel protein-protein interaction inhibitors of the PRC2 complex through EED binding.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
6KEI
 
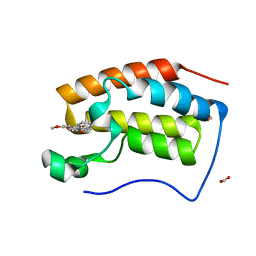 | | Crystal structure of BRD4 bromodomain 1 (BD1) in complex with 16-methoxy-11-methyl-6-[(pyridin-2-yl)methoxy]-2-oxa-11-azatetracyclo[8.6.1.03,8.013,17]heptadeca-1(16),3,5,7,9,13(17),14-heptaen-12-one | | Descriptor: | 16-methoxy-11-methyl-6-[(pyridin-2-yl)methoxy]-2-oxa-11-azatetracyclo[8.6.1.03,8.013,17]heptadeca-1(16),3,5,7,9,13(17),14-heptaen-12-one, Bromodomain-containing protein 4, FORMIC ACID | | Authors: | Lee, B.I, Park, T.H. | | Deposit date: | 2019-07-04 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | Synthesis and Structure-Activity Relationships of Aristoyagonine Derivatives as Brd4 Bromodomain Inhibitors with X-ray Co-Crystal Research.
Molecules, 26, 2021
|
|
