6IPZ
 
 | | Fyn SH3 domain R96W mutant, crystallized with 18-crown-6 | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, Tyrosine-protein kinase Fyn | | Authors: | Arold, S.T, Aljedani, S.S, Shahul Hameed, U.F. | | Deposit date: | 2018-11-05 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.576 Å) | | Cite: | Synergy and allostery in ligand binding by HIV-1 Nef.
Biochem.J., 478, 2021
|
|
3GHH
 
 | | Structural insights into the catalytic mechanism of CD38: Evidence for a conformationally flexible covalent enzyme-substrate complex. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ecto-NAD+ glycohydrolase (CD38 molecule), SULFATE ION, ... | | Authors: | Egea, P.F, Muller-Steffner, H, Stroud, R.M, Oppenheimer, N.J, Kellenberger, E, Schuber, F. | | Deposit date: | 2009-03-03 | | Release date: | 2010-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Insights into the mechanism of bovine CD38/NAD+glycohydrolase from the X-ray structures of its Michaelis complex and covalently-trapped intermediates.
Plos One, 7, 2012
|
|
6J2A
 
 | | The structure of HLA-A*3003/NP44 | | Descriptor: | Beta-2-microglobulin, HLA-A*3003, NP44 | | Authors: | Zhu, S.Y, Liu, K.F, Chai, Y, Ding, C.M, Lv, J.X, Gao, F.G, Lou, Y.L, Liu, W.J. | | Deposit date: | 2018-12-31 | | Release date: | 2019-09-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Divergent Peptide Presentations of HLA-A*30 Alleles Revealed by Structures With Pathogen Peptides.
Front Immunol, 10, 2019
|
|
6J1W
 
 | | The structure of HLA-A*3001/RT313 | | Descriptor: | ALA-ILE-PHE-GLN-SER-SER-MET-THR-LYS, Beta-2-microglobulin, HLA-A*3001 | | Authors: | Zhu, S.Y, Liu, K.F, Chai, Y, Ding, C.M, Lv, J.X, Gao, G.F, Lou, Y.L, Liu, W.J. | | Deposit date: | 2018-12-29 | | Release date: | 2019-09-25 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Divergent Peptide Presentations of HLA-A*30 Alleles Revealed by Structures With Pathogen Peptides.
Front Immunol, 10, 2019
|
|
13PK
 
 | | TERNARY COMPLEX OF PHOSPHOGLYCERATE KINASE FROM TRYPANOSOMA BRUCEI | | Descriptor: | 3-PHOSPHOGLYCERATE KINASE, 3-PHOSPHOGLYCERIC ACID, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Bernstein, B.E, Michels, P.A.M, Hol, W.G.J. | | Deposit date: | 1996-11-23 | | Release date: | 1997-12-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Synergistic effects of substrate-induced conformational changes in phosphoglycerate kinase activation.
Nature, 385, 1997
|
|
1ADJ
 
 | | HISTIDYL-TRNA SYNTHETASE IN COMPLEX WITH HISTIDINE | | Descriptor: | HISTIDINE, HISTIDYL-TRNA SYNTHETASE, SULFATE ION | | Authors: | Cusack, S, Aberg, A. | | Deposit date: | 1997-02-18 | | Release date: | 1997-08-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure analysis of the activation of histidine by Thermus thermophilus histidyl-tRNA synthetase.
Biochemistry, 36, 1997
|
|
6J29
 
 | | The structure of HLA-A*3003/MTB | | Descriptor: | Beta-2-microglobulin, HLA-A*3003, MTB | | Authors: | Zhu, S.Y, Liu, K.F, Chai, Y, Ding, C.M, Lv, J.X, Gao, F.G, Lou, Y.L, Liu, W.J. | | Deposit date: | 2018-12-31 | | Release date: | 2019-09-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Divergent Peptide Presentations of HLA-A*30 Alleles Revealed by Structures With Pathogen Peptides.
Front Immunol, 10, 2019
|
|
8CBD
 
 | | SARS-CoV-2 Delta-RBD complexed with BA.4/5-1 and EY6A Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BA.4/5-1 heavy chain, BA.4/5-1 light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2023-01-25 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.52 Å) | | Cite: | Emerging variants develop total escape from potent monoclonal antibodies induced by BA.4/5 infection.
Nat Commun, 15, 2024
|
|
5MYJ
 
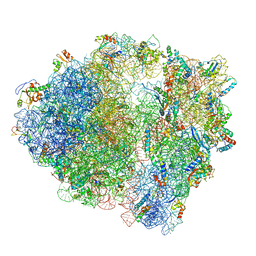 | | Structure of 70S ribosome from Lactococcus lactis | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Franken, L.E, Oostergetel, G.T, Pijning, T, Puri, P, Boekema, E.J, Poolman, B, Guskov, A. | | Deposit date: | 2017-01-26 | | Release date: | 2017-10-11 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | A general mechanism of ribosome dimerization revealed by single-particle cryo-electron microscopy.
Nat Commun, 8, 2017
|
|
8CBF
 
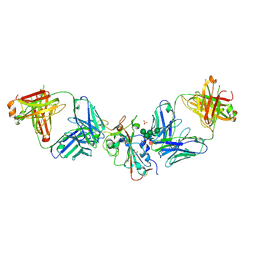 | | SARS-CoV-2 Delta-RBD complexed with Omi-42 and Beta-49 Fabs | | Descriptor: | Beta-49 heavy chain, Beta-49 light chain, CHLORIDE ION, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2023-01-25 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Emerging variants develop total escape from potent monoclonal antibodies induced by BA.4/5 infection.
Nat Commun, 15, 2024
|
|
8CBE
 
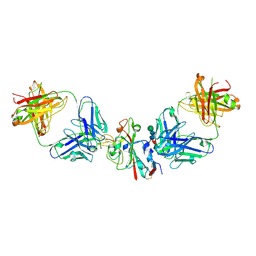 | | SARS-CoV-2 Delta-RBD complexed with BA.4/5-2 and Beta-49 Fabs | | Descriptor: | BA.4/5-2 heavy chain, BA.4/5-2 light chain, Beta-49 heavy chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2023-01-25 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Emerging variants develop total escape from potent monoclonal antibodies induced by BA.4/5 infection.
Nat Commun, 15, 2024
|
|
6J1V
 
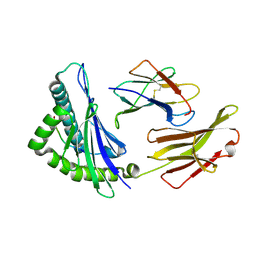 | | The structure of HLA-A*3003/RT313 | | Descriptor: | Beta-2-microglobulin, HLA-A*3003, RT313 | | Authors: | Zhu, S.Y, Liu, K.F, Chai, Y, Ding, C.M, Lv, J.X, Gao, G.F, Lou, Y.L, Liu, W.J. | | Deposit date: | 2018-12-29 | | Release date: | 2019-09-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Divergent Peptide Presentations of HLA-A*30 Alleles Revealed by Structures With Pathogen Peptides.
Front Immunol, 10, 2019
|
|
8CMA
 
 | | SARS-CoV-2 Delta-RBD complexed with BA.4/5-35 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BA.4/5-35 heavy chain, BA.4/5-35 light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2023-02-18 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Emerging variants develop total escape from potent monoclonal antibodies induced by BA.4/5 infection.
Nat Commun, 15, 2024
|
|
2HL6
 
 | | Structure of homologously expressed Ferrulate esterase of Aspergillus niger in complex with CAPS | | Descriptor: | 1,2-ETHANEDIOL, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, Feruloyl esterase A, ... | | Authors: | Parsiegla, G, Herpoel-Gimbert, I, Dubots, J. | | Deposit date: | 2006-07-06 | | Release date: | 2006-10-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Respective importance of protein folding and glycosylation in the thermal stability of recombinant feruloyl esterase A
Febs Lett., 580, 2006
|
|
1A06
 
 | | CALMODULIN-DEPENDENT PROTEIN KINASE FROM RAT | | Descriptor: | CALCIUM/CALMODULIN-DEPENDENT PROTEIN KINASE | | Authors: | Kuriyan, J, Goldberg, J. | | Deposit date: | 1997-12-09 | | Release date: | 1998-04-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the autoinhibition of calcium/calmodulin-dependent protein kinase I.
Cell(Cambridge,Mass.), 84, 1996
|
|
1EC0
 
 | | HIV-1 protease in complex with the inhibitor bea403 | | Descriptor: | HIV-1 PROTEASE, N,N-[2,5-O-DI-2-FLUORO-BENZYL-GLUCARYL]-DI-[1-AMINO-INDAN-2-OL] | | Authors: | Unge, T. | | Deposit date: | 2000-01-25 | | Release date: | 2002-06-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Symmetric fluoro-substituted diol-based HIV protease inhibitors. Ortho-fluorinated and meta-fluorinated P1/P1'-benzyloxy side groups significantly improve the antiviral activity and preserve binding efficacy
Eur.J.Biochem., 271, 2004
|
|
1ATE
 
 | |
1ATA
 
 | |
5NUP
 
 | | Structural basis for maintenance of bacterial outer membrane lipid asymmetry | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, ABC transporter permease, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Abellon-Ruiz, J, Kaptan, S.S, Basle, A, Claudi, B, Bumann, D, Kleinekathofer, U, van den Berg, B. | | Deposit date: | 2017-05-01 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for maintenance of bacterial outer membrane lipid asymmetry.
Nat Microbiol, 2, 2017
|
|
5NUQ
 
 | | Structural basis for maintenance of bacterial outer membrane lipid asymmetry | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, Outer membrane protein F, Probable phospholipid-binding lipoprotein mlaA | | Authors: | Abellon-Ruiz, J, Kaptan, S.S, Basle, A, Claudi, B, Bumann, D, Kleinekathofer, U, van den Berg, B. | | Deposit date: | 2017-05-01 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for maintenance of bacterial outer membrane lipid asymmetry.
Nat Microbiol, 2, 2017
|
|
5NUR
 
 | | Structural basis for maintenance of bacterial outer membrane lipid asymmetry | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-4)-3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-6)-2-amino-2-deoxy-4-O-phosphono-beta-D-glucopyranose-(1-6)-2-amino-2-deoxy-1-O-phosphono-alpha-D-glucopyranose, 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(3-4)-3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-6)-2-amino-2-deoxy-4-O-phosphono-beta-D-glucopyranose-(1-6)-2-amino-2-deoxy-1-O-phosphono-alpha-D-glucopyranose, ... | | Authors: | Abellon-Ruiz, J, Kaptan, S.S, Basle, A, Claudi, B, Bumann, D, Kleinekathofer, U, van den Berg, B. | | Deposit date: | 2017-05-01 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Structural basis for maintenance of bacterial outer membrane lipid asymmetry.
Nat Microbiol, 2, 2017
|
|
5NUO
 
 | | Structural basis for maintenance of bacterial outer membrane lipid asymmetry | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, ABC transporter permease, Outer membrane protein F, ... | | Authors: | Abellon-Ruiz, J, Kaptan, S.S, Basle, A, Claudi, B, Bumann, D, Kleinekathofer, U, van den Berg, B. | | Deposit date: | 2017-05-01 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for maintenance of bacterial outer membrane lipid asymmetry.
Nat Microbiol, 2, 2017
|
|
3IBP
 
 | | The Crystal Structure of the Dimerization Domain of Escherichia coli Structural Maintenance of Chromosomes Protein MukB | | Descriptor: | AMMONIUM ION, Chromosome partition protein mukB | | Authors: | Li, Y, Schoeffler, A.J, Berger, J.M, Oakley, M.G. | | Deposit date: | 2009-07-16 | | Release date: | 2010-01-26 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (3.099 Å) | | Cite: | The crystal structure of the hinge domain of the Escherichia coli structural maintenance of chromosomes protein MukB.
J.Mol.Biol., 395, 2010
|
|
5HVY
 
 | | CDK8/CYCC IN COMPLEX WITH COMPOUND 20 | | Descriptor: | CHLORIDE ION, Cyclin-C, Cyclin-dependent kinase 8, ... | | Authors: | Kiefer, J.R, Schneider, E.V, Maskos, K, Bergeron, P, Koehler, M. | | Deposit date: | 2016-01-28 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Design and Development of a Series of Potent and Selective Type II Inhibitors of CDK8.
Acs Med.Chem.Lett., 7, 2016
|
|
1ATB
 
 | |
