6EBG
 
 | | Ohr (Organic Hydroperoxide Resistance protein) mutant - C60S interacting with dihydrolipoamide | | Descriptor: | (6S)-6,8-disulfanyloctanamide, Organic hydroperoxide resistance protein | | Authors: | Domingos, R.M, Teixeira, R.D, Alegria, T.G.P, Vieira, P.S, Murakami, M.T, Netto, L.E.S. | | Deposit date: | 2018-08-06 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Substrate and product-assisted catalysis: molecular aspects behind structural switches along Organic Hydroperoxide Resistance Protein catalytic cycle
Acs Catalysis, 2020
|
|
6EBT
 
 | |
6ECD
 
 | | Vlm2 thioesterase domain with genetically encoded 2,3-diaminopropionic acid bound with a tetradepsipeptide | | Descriptor: | Vlm2, tetradepsipeptide | | Authors: | Alonzo, D.A, Huguenin-Dezot, N, Heberlig, G.W, Mahesh, M, Nguyen, D.P, Dornan, M.H, Boddy, C.N, Chin, J.W, Schmeing, T.M. | | Deposit date: | 2018-08-07 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Trapping biosynthetic acyl-enzyme intermediates with encoded 2,3-diaminopropionic acid.
Nature, 565, 2019
|
|
6KDO
 
 | | HIV-1 reverse transcriptase with Q151M/Y115F/F116Y/M184V/F160M:DNA:lamivudine 5'-triphosphate ternary complex | | Descriptor: | DNA/RNA (38-MER), GLYCEROL, HIV-1 RT p51 subunit, ... | | Authors: | Yasutake, Y, Hattori, S.I, Tamura, N, Maeda, K. | | Deposit date: | 2019-07-02 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.573 Å) | | Cite: | Structural features in common of HBV and HIV-1 resistance against chirally-distinct nucleoside analogues entecavir and lamivudine.
Sci Rep, 10, 2020
|
|
6EEB
 
 | | Calmodulin in complex with malbrancheamide | | Descriptor: | (5aS,12aS,13aS)-8,9-dichloro-12,12-dimethyl-2,3,11,12,12a,13-hexahydro-1H,5H,6H-5a,13a-(epiminomethano)indolizino[7,6-b]carbazol-14-one, CALCIUM ION, Calmodulin-1, ... | | Authors: | Beyett, T.S, Fraley, A.E, Tesmer, J.J.G. | | Deposit date: | 2018-08-13 | | Release date: | 2019-08-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Perturbation of the interactions of calmodulin with GRK5 using a natural product chemical probe.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6EDK
 
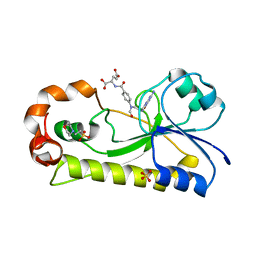 | | Crystal structure of the formyltransferase PseJ from Anoxybacillus kamchatkensis with N10-formyltetrahydrofolate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Formyltransferase PseJ, N-{4-[{[(6S)-2-amino-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}(formyl)amino]benzoyl}-L-glutamic acid, ... | | Authors: | Reimer, J.M, Harb, I, Schmeing, T.M. | | Deposit date: | 2018-08-09 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insight into a Novel Formyltransferase and Evolution to a Nonribosomal Peptide Synthetase Tailoring Domain.
ACS Chem. Biol., 13, 2018
|
|
6KEN
 
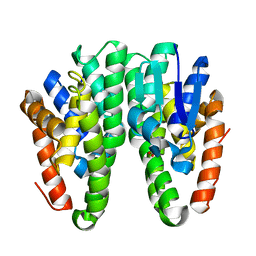 | | Crystal structure of Drosophila melanogaster Noppera-bo, glutathione S-transferase epsilon 14 (DmGSTE14), in glutathione-bound form | | Descriptor: | GLUTATHIONE, Glutathione S-transferase E14 | | Authors: | Koiwai, K, Inaba, K, Morohashi, K, Yumoto, F, Niwa, R, Senda, T. | | Deposit date: | 2019-07-04 | | Release date: | 2019-10-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | An integrated approach to unravel a crucial structural property required for the function of the insect steroidogenic Halloween protein Noppera-bo.
J.Biol.Chem., 295, 2020
|
|
6KGK
 
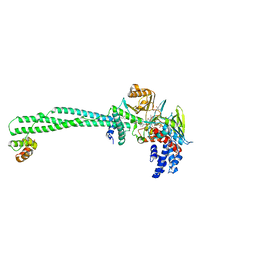 | | LSD1-CoREST-S2101 five-membered ring adduct model | | Descriptor: | 3-[3,5-bis(fluoranyl)-2-phenylmethoxy-phenyl]propanal, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Niwa, H, Sato, S, Sengoku, S, Umehara, T, Yokoyama, S. | | Deposit date: | 2019-07-12 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Development and Structural Evaluation of N-Alkylated trans-2-Phenylcyclopropylamine-Based LSD1 Inhibitors.
Chemmedchem, 15, 2020
|
|
6EFG
 
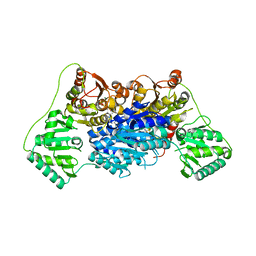 | | Pyruvate decarboxylase from Kluyveromyces lactis | | Descriptor: | MAGNESIUM ION, Pyruvate decarboxylase, THIAMINE DIPHOSPHATE | | Authors: | Kutter, S, Konig, S. | | Deposit date: | 2018-08-16 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | The crystal structures of pyruvate decarboxylase from Kluyveromyces lactis in the absence of ligands and in the presence of the substrate surrogate pyruvamide
To be Published
|
|
6EHH
 
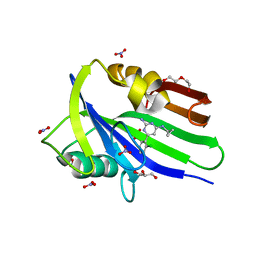 | | Crystal structure of mouse MTH1 mutant L116M with inhibitor TH588 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, COPPER (II) ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gustafsson, R, Narwal, M, Jemth, A.-S, Almlof, I, Warpman Berglund, U, Helleday, T, Stenmark, P. | | Deposit date: | 2017-09-13 | | Release date: | 2018-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures and Inhibitor Interactions of Mouse and Dog MTH1 Reveal Species-Specific Differences in Affinity.
Biochemistry, 57, 2018
|
|
6K24
 
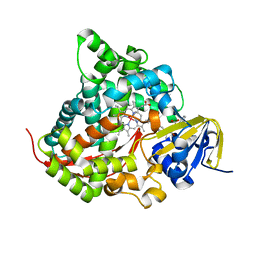 | | Structure of the Rhodium Mesoporphyrin IX-Reconstituted CYP102A1 Haem Domain with N-Abietoyl-L-Tryptophan | | Descriptor: | (2S)-2-[[(1R,4aR,4bR,10aR)-1,4a-dimethyl-7-propan-2-yl-2,3,4,4b,5,6,10,10a-octahydrophenanthren-1-yl]carbonylamino]-3-( 1H-indol-3-yl)propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, DIMETHYL SULFOXIDE, ... | | Authors: | Stanfield, J.K, Matsumoto, A, Kasai, C, Sugimoto, H, Shiro, Y, Watanabe, Y, Shoji, O. | | Deposit date: | 2019-05-13 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystals in Minutes: Instant On-Site Microcrystallisation of Various Flavours of the CYP102A1 (P450BM3) Haem Domain.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6EI0
 
 | | Cytosolic copper storage protein Csp from Streptomyces lividans: apo form | | Descriptor: | Cytosolic copper storage protein (Ccsp), GLYCEROL, SULFATE ION, ... | | Authors: | Straw, M.L, Chaplin, A.K, Hough, M.A, Worrall, J.A.R. | | Deposit date: | 2017-09-15 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | A cytosolic copper storage protein provides a second level of copper tolerance in Streptomyces lividans.
Metallomics, 10, 2018
|
|
6EG0
 
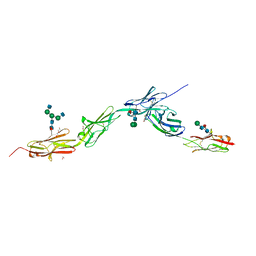 | | Crystal structure of Dpr4 Ig1-Ig2 in complex with DIP-Eta Ig1-Ig3 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cosmanescu, F, Shapiro, L. | | Deposit date: | 2018-08-17 | | Release date: | 2018-11-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Neuron-Subtype-Specific Expression, Interaction Affinities, and Specificity Determinants of DIP/Dpr Cell Recognition Proteins.
Neuron, 100, 2018
|
|
6KJJ
 
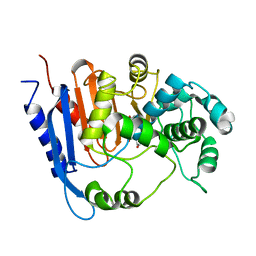 | | Functional and structural insights into the unusual oxyanion hole-like geometry in macrolactin acyltransferase selective for dicarboxylic acyl donors | | Descriptor: | 4-(2-acetamidoethylsulfanyl)-4-oxidanylidene-butanoic acid, Putative beta-lactamase | | Authors: | Xiao, F, Dong, S, Feng, Y, Li, W. | | Deposit date: | 2019-07-22 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.492 Å) | | Cite: | Structural Basis of Specificity for Carboxyl-Terminated Acyl Donors in a Bacterial Acyltransferase.
J.Am.Chem.Soc., 142, 2020
|
|
6KK6
 
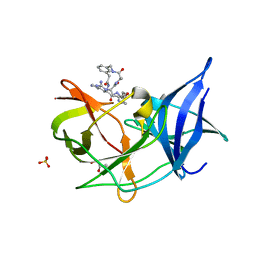 | | Crystal structure of Zika NS2B-NS3 protease with compound 16 | | Descriptor: | 1-[(5~{R},8~{R},15~{S},18~{S})-15,18-bis(4-azanylbutyl)-5-methyl-4,7,14,17,20-pentakis(oxidanylidene)-3,6,13,16,19-pentazabicyclo[20.3.1]hexacosa-1(25),22(26),23-trien-8-yl]guanidine, GLYCEROL, NS3 protease, ... | | Authors: | Quek, J.P. | | Deposit date: | 2019-07-23 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structure-Based Macrocyclization of Substrate Analogue NS2B-NS3 Protease Inhibitors of Zika, West Nile and Dengue viruses.
Chemmedchem, 15, 2020
|
|
6EJD
 
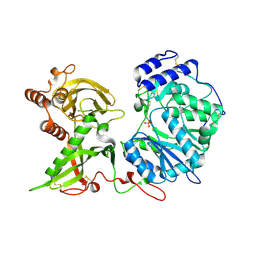 | |
6EKH
 
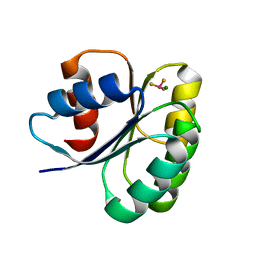 | |
6EKZ
 
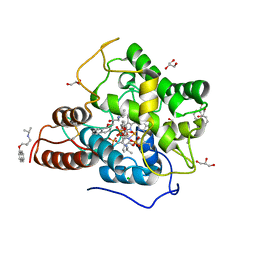 | |
6EH6
 
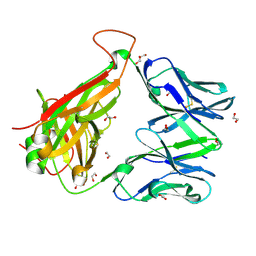 | |
6EI3
 
 | | Crystal structure of auto inhibited POT family peptide transporter | | Descriptor: | (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, Proton-dependent oligopeptide transporter family protein | | Authors: | Newstead, S, Brinth, A, Vogeley, L, Caffrey, M. | | Deposit date: | 2017-09-17 | | Release date: | 2017-11-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Proton movement and coupling in the POT family of peptide transporters.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6EII
 
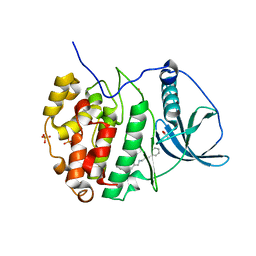 | | The crystal structure of CK2alpha in complex with compound 18 | | Descriptor: | (3-chloranyl-4-phenyl-phenyl)methyl-(3-phenylpropyl)azanium, ACETATE ION, Casein kinase II subunit alpha, ... | | Authors: | Brear, P, De Fusco, C, Iegre, J, Yoshida, M, Mitchell, S, Rossmann, M, Carro, L, Sore, H, Hyvonen, M, Spring, D. | | Deposit date: | 2017-09-19 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.935 Å) | | Cite: | Second-generation CK2 alpha inhibitors targeting the alpha D pocket.
Chem Sci, 9, 2018
|
|
6K7Q
 
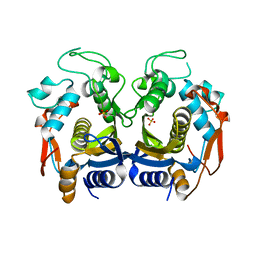 | | Crystal structure of thymidylate synthase from shrimp | | Descriptor: | SULFATE ION, Thymidylate synthase | | Authors: | Ma, Q, Zang, K, Liu, C. | | Deposit date: | 2019-06-08 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural analysis of a shrimp thymidylate synthase reveals species-specific interactions with dUMP and raltitrexed.
J Oceanol Limnol, 38, 2020
|
|
6EIZ
 
 | | A de novo designed hexameric coiled coil CC-Hex2 with farnesol bound in the channel. | | Descriptor: | (2E,6E)-3,7,11-trimethyldodeca-2,6,10-trien-1-ol, CC-Hex2 | | Authors: | Rhys, G.G, Burton, A.J, Dawson, W.M, Thomas, F, Woolfson, D.N. | | Deposit date: | 2017-09-19 | | Release date: | 2018-07-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | De Novo-Designed alpha-Helical Barrels as Receptors for Small Molecules.
ACS Synth Biol, 7, 2018
|
|
6KAY
 
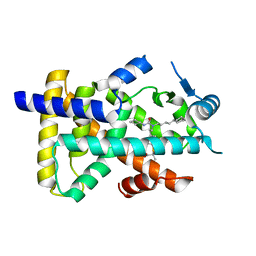 | | X-ray structure of human PPARalpha ligand binding domain-GW7647 co-crystals obtained by soaking | | Descriptor: | 2-[(4-{2-[(4-cyclohexylbutyl)(cyclohexylcarbamoyl)amino]ethyl}phenyl)sulfanyl]-2-methylpropanoic acid, GLYCEROL, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Suda, K, Saito, K, Oyama, T, Ishii, I. | | Deposit date: | 2019-06-24 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.735 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6KB3
 
 | | X-ray structure of human PPARalpha ligand binding domain-GW7647 co-crystals obtained by delipidation and cross-seeding | | Descriptor: | 2-[(4-{2-[(4-cyclohexylbutyl)(cyclohexylcarbamoyl)amino]ethyl}phenyl)sulfanyl]-2-methylpropanoic acid, GLYCEROL, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2019-06-24 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
