2QLV
 
 | |
2QH1
 
 | | Structure of TA289, a CBS-rubredoxin-like protein, in its Fe+2-bound state | | Descriptor: | FE (II) ION, Hypothetical protein Ta0289 | | Authors: | Singer, A.U, Proudfoot, M, Brown, G, Xu, L, Savchenko, A, Yakunin, A.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-06-29 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemical and structural characterization of a novel family of cystathionine beta-synthase domain proteins fused to a Zn ribbon-like domain.
J.Mol.Biol., 375, 2008
|
|
2PFI
 
 | |
2P9M
 
 | | Crystal structure of conserved hypothetical protein MJ0922 from Methanocaldococcus jannaschii DSM 2661 | | Descriptor: | Hypothetical protein MJ0922 | | Authors: | Zhao, M, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, Zhu, J, Swindell II, J.T, Chen, L, Fu, Z.-Q, Charz, J, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-26 | | Release date: | 2007-07-03 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structure of conserved hypothetical protein MJ0922 from Methanocaldococcus jannaschii DSM 2661
To be Published
|
|
2OUX
 
 | |
2OOY
 
 | |
2OOX
 
 | |
2O16
 
 | | Crystal structure of a putative acetoin utilization protein (AcuB) from Vibrio cholerae | | Descriptor: | Acetoin utilization protein AcuB, putative, PHOSPHATE ION | | Authors: | Patskovsky, Y, Bonanno, J.B, Rutter, M, Bain, K.T, Powell, A, Slocombe, A, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-11-28 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a putative acetoin utilization protein (AcuB) from Vibrio cholerae
To be Published
|
|
2NYE
 
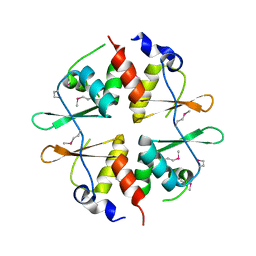 | | Crystal structure of the Bateman2 domain of yeast Snf4 | | Descriptor: | Nuclear protein SNF4 | | Authors: | Rudolph, M.J, Amodeo, G.A, Iram, S, Hong, S, Pirino, G, Carlson, M, Tong, L. | | Deposit date: | 2006-11-20 | | Release date: | 2006-12-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Bateman2 domain of yeast Snf4: dimeric association and relevance for AMP binding.
Structure, 15, 2007
|
|
2NYC
 
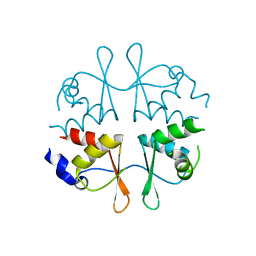 | | Crystal structure of the Bateman2 domain of yeast Snf4 | | Descriptor: | Nuclear protein SNF4 | | Authors: | Rudolph, M.J, Amodeo, G.A, Iram, S, Hong, S, Pirino, G, Carlson, M, Tong, L. | | Deposit date: | 2006-11-20 | | Release date: | 2006-12-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the Bateman2 domain of yeast Snf4: dimeric association and relevance for AMP binding.
Structure, 15, 2007
|
|
2JA3
 
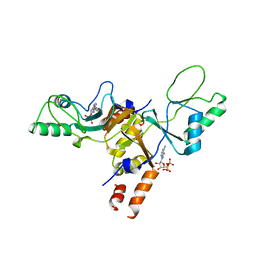 | | Cytoplasmic Domain of the Human Chloride Transporter ClC-5 in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE CHANNEL PROTEIN 5 | | Authors: | Meyer, S, Savaresi, S, Forster, I.C, Dutzler, R. | | Deposit date: | 2006-11-21 | | Release date: | 2007-01-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Nucleotide Recognition by the Cytoplasmic Domain of the Human Chloride Transporter Clc-5
Nat.Struct.Mol.Biol., 14, 2006
|
|
2J9L
 
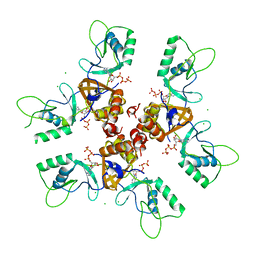 | | Cytoplasmic Domain of the Human Chloride Transporter ClC-5 in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE CHANNEL PROTEIN 5, CHLORIDE ION | | Authors: | Meyer, S, Savaresi, S, Forster, I.C, Dutzler, R. | | Deposit date: | 2006-11-13 | | Release date: | 2007-01-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Nucleotide Recognition by the Cytoplasmic Domain of the Human Chloride Transporter Clc-5
Nat.Struct.Mol.Biol., 14, 2006
|
|
2EMQ
 
 | |
2EF7
 
 | | Crystal structure of ST2348, a hypothetical protein with CBS domains from Sulfolobus tokodaii strain7 | | Descriptor: | Hypothetical protein ST2348 | | Authors: | Agari, Y, Karthe, P, Kumarevel, T, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-21 | | Release date: | 2007-08-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of ST2348, a CBS domain protein, from hyperthermophilic archaeon Sulfolobus tokodaii
Biochem.Biophys.Res.Commun., 375, 2008
|
|
2D4Z
 
 | |
2CU0
 
 | |
1ZFJ
 
 | | INOSINE MONOPHOSPHATE DEHYDROGENASE (IMPDH; EC 1.1.1.205) FROM STREPTOCOCCUS PYOGENES | | Descriptor: | INOSINE MONOPHOSPHATE DEHYDROGENASE, INOSINIC ACID | | Authors: | Zhang, R, Evans, G, Rotella, F.J, Westbrook, E.M, Beno, D, Huberman, E, Joachimiak, A, Collart, F.R. | | Deposit date: | 1999-03-29 | | Release date: | 2000-03-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characteristics and crystal structure of bacterial inosine-5'-monophosphate dehydrogenase.
Biochemistry, 38, 1999
|
|
1Y5H
 
 | |
1XKF
 
 | |
1VRD
 
 | |
1VR9
 
 | |
1PVM
 
 | | Crystal Structure of a Conserved CBS Domain Protein TA0289 of Unknown Function from Thermoplasma acidophilum | | Descriptor: | MERCURY (II) ION, conserved hypothetical protein Ta0289 | | Authors: | Zhang, R, Joachimiak, A, Edwards, A, Savchenko, A, Xu, L, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-06-27 | | Release date: | 2004-01-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Biochemical and structural characterization of a novel family of cystathionine beta-synthase domain proteins fused to a Zn ribbon-like domain
J.Mol.Biol., 375, 2008
|
|
1PBJ
 
 | | CBS domain protein | | Descriptor: | MAGNESIUM ION, hypothetical protein | | Authors: | Cuff, M.E, Skarina, T, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-05-14 | | Release date: | 2003-12-16 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of a hypothetical protein from M. thermautotrophicus reveals a
novel fold and a pseudo 2-fold axis of symmetry
TO BE PUBLISHED
|
|
1O50
 
 | |
1NFB
 
 | |
