2G0B
 
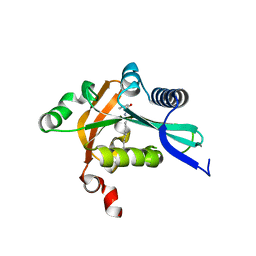 | | The structure of FeeM, an N-acyl amino acid synthase from uncultured soil microbes | | Descriptor: | FeeM, N-DODECANOYL-L-TYROSINE | | Authors: | Van Wagoner, R.M, Clardy, J. | | Deposit date: | 2006-02-11 | | Release date: | 2006-09-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | FeeM, an N-Acyl Amino Acid Synthase from an Uncultured Soil Microbe: Structure, Mechanism, and Acyl Carrier Protein Binding.
Structure, 14, 2006
|
|
5T7D
 
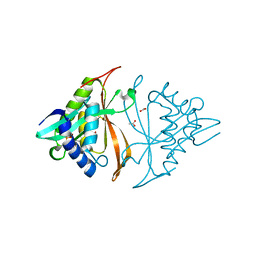 | |
2EVN
 
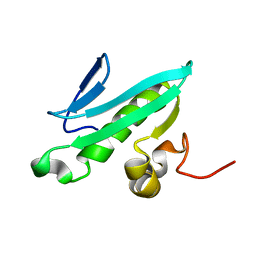 | | NMR solution structures of At1g77540 | | Descriptor: | Protein At1g77540 | | Authors: | Tyler, R.C, Singh, S, Tonelli, M, Min, M.S, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-10-31 | | Release date: | 2005-11-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of Arabidopsis thaliana At1g77540 Protein, a Minimal Acetyltransferase from the COG2388 Family.
Biochemistry, 45, 2006
|
|
5T7E
 
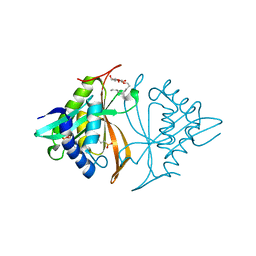 | |
2DPS
 
 | |
5GI6
 
 | |
6F3T
 
 | | Crystal structure of the human TAF5-TAF6-TAF9 complex | | Descriptor: | CHLORIDE ION, Transcription initiation factor TFIID subunit 5, Transcription initiation factor TFIID subunit 6, ... | | Authors: | Haffke, M, Berger, I. | | Deposit date: | 2017-11-28 | | Release date: | 2018-12-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Chaperonin CCT checkpoint function in basal transcription factor TFIID assembly.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5GIG
 
 | | Crystal Structure of Drosophila melanogaster E47D Dopamine N-Acetyltransferase in Ternary Complex with CoA and Acetyl-dopamine | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, COENZYME A, Dopamine N-acetyltransferase, ... | | Authors: | Yang, Y.C, Wu, C.Y, Cheng, H.C, Lyu, P.C. | | Deposit date: | 2016-06-23 | | Release date: | 2017-07-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structure of Drosophila melanogaster E47D Dopamine N-Acetyltransferase in Ternary Complex with CoA and Acetyl-dopamine
To Be Published
|
|
5GII
 
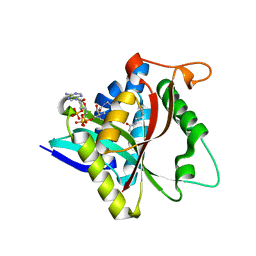 | |
5GI8
 
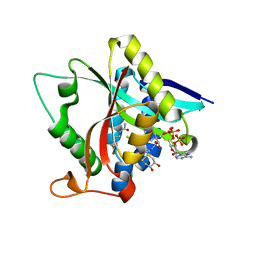 | | Crystal Structure of Drosophila melanogaster Dopamine N-Acetyltransferase in Ternary Complex with CoA and Acetyl-dopamine | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, COENZYME A, Dopamine N-acetyltransferase, ... | | Authors: | Yang, Y.C, Lin, S.J, Cheng, K.C, Cheng, H.C, Lyu, P.C. | | Deposit date: | 2016-06-22 | | Release date: | 2017-07-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structure of Drosophila melanogaster Dopamine N-Acetyltransferase in Ternary Complex with CoA and Acetyl-dopamine
To Be Published
|
|
5HMN
 
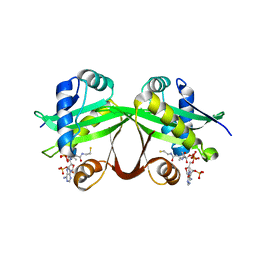 | | Crystal structure of an aminoglycoside acetyltransferase HMB0005 from an uncultured soil metagenomic sample, unknown active site density modeled as polyethylene glycol | | Descriptor: | AAC3-I, COENZYME A, TETRAETHYLENE GLYCOL | | Authors: | Xu, Z, Stogios, P.J, Wawrzak, Z, Skarina, T, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-16 | | Release date: | 2016-02-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.018 Å) | | Cite: | Crystal structure of an aminoglycoside acetyltransferase HMB0005 from an uncultured soil metagenomic sample, unknown active site density modeled as polyethylene glycol
To Be Published
|
|
7XRJ
 
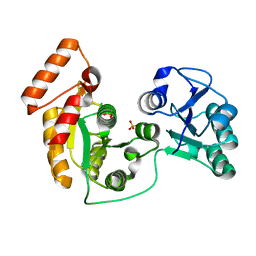 | | crystal structure of N-acetyltransferase DgcN-25328 | | Descriptor: | Putative NAD-dependent epimerase/dehydratase family protein, SULFATE ION | | Authors: | Zhang, Y.Z, Yu, Y, Cao, H.Y, Chen, X.L, Wang, P. | | Deposit date: | 2022-05-10 | | Release date: | 2023-02-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Novel D-glutamate catabolic pathway in marine Proteobacteria and halophilic archaea.
Isme J, 17, 2023
|
|
1I12
 
 | |
1I1D
 
 | | CRYSTAL STRUCTURE OF YEAST GNA1 BOUND TO COA AND GLNAC-6P | | Descriptor: | 2-acetamido-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, COENZYME A, GLUCOSAMINE-PHOSPHATE N-ACETYLTRANSFERASE, ... | | Authors: | Peneff, C, Mengin-Lecreulx, D, Bourne, Y. | | Deposit date: | 2001-02-01 | | Release date: | 2001-05-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structures of Apo and complexed Saccharomyces cerevisiae GNA1 shed light on the catalytic mechanism of an amino-sugar N-acetyltransferase.
J.Biol.Chem., 276, 2001
|
|
1J4J
 
 | | Crystal Structure of Tabtoxin Resistance Protein (form II) complexed with an Acyl Coenzyme A | | Descriptor: | ACETYL COENZYME *A, TABTOXIN RESISTANCE PROTEIN | | Authors: | He, H, Ding, Y, Bartlam, M, Zhang, R, Duke, N, Joachimiak, A, Shao, Y, Cao, Z, Tang, H, Liu, Y, Jiang, F, Liu, J, Zhao, N, Rao, Z. | | Deposit date: | 2001-10-02 | | Release date: | 2003-06-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of tabtoxin resistance protein complexed with acetyl coenzyme A reveals the mechanism for beta-lactam acetylation.
J.Mol.Biol., 325, 2003
|
|
1I21
 
 | | CRYSTAL STRUCTURE OF YEAST GNA1 | | Descriptor: | GLUCOSAMINE-PHOSPHATE N-ACETYLTRANSFERASE | | Authors: | Peneff, C, Mengin-Lecreulx, D, Bourne, Y. | | Deposit date: | 2001-02-05 | | Release date: | 2001-05-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structures of Apo and complexed Saccharomyces cerevisiae GNA1 shed light on the catalytic mechanism of an amino-sugar N-acetyltransferase.
J.Biol.Chem., 276, 2001
|
|
6CY6
 
 | | Crystal structure of spermidine/spermine N-acetyltransferase SpeG from Escherichia coli in complex with tris(hydroxymethyl)aminomethane. | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-04-04 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Analysis of crystalline and solution states of ligand-free spermidine N-acetyltransferase (SpeG) from Escherichia coli.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
8Q6U
 
 | |
8GXJ
 
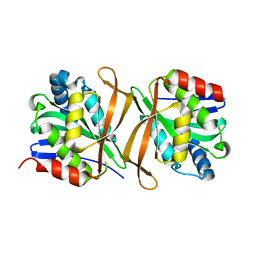 | |
8GXK
 
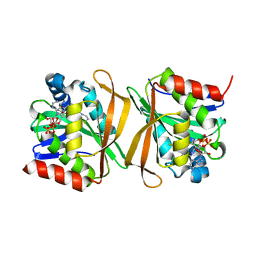 | | Pseudomonas jinjuensis N-acetyltransferase | | Descriptor: | COENZYME A, Protein N-acetyltransferase, RimJ/RimL family | | Authors: | Song, Y.J, Bao, R. | | Deposit date: | 2022-09-20 | | Release date: | 2022-10-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The novel type II toxin-antitoxin PacTA modulates Pseudomonas aeruginosa iron homeostasis by obstructing the DNA-binding activity of Fur.
Nucleic Acids Res., 50, 2022
|
|
8Q26
 
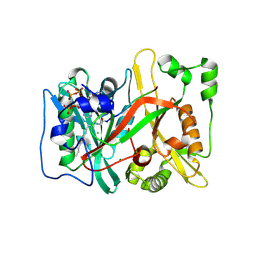 | | HsNMT1 in complex with both MyrCoA and GNCFSKPR inhibitor peptide | | Descriptor: | CHLORIDE ION, COENZYME A, Glycylpeptide N-tetradecanoyltransferase 1, ... | | Authors: | Dian, C, Giglione, C, Meinnel, T. | | Deposit date: | 2023-08-01 | | Release date: | 2024-08-14 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Novel, tightly structurally related N-myristoyltransferase inhibitors display equally potent yet distinct inhibitory mechanisms.
Structure, 32, 2024
|
|
8Q3D
 
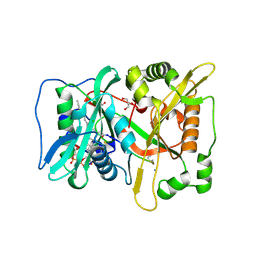 | | HsNMT1 in complex with both MyrCoA and GNCFSKPR(NH2) inhibitor peptide | | Descriptor: | CHLORIDE ION, COENZYME A, GLYCEROL, ... | | Authors: | Dian, C, Giglione, C, Meinnel, T. | | Deposit date: | 2023-08-04 | | Release date: | 2024-08-14 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Novel, tightly structurally related N-myristoyltransferase inhibitors display equally potent yet distinct inhibitory mechanisms.
Structure, 32, 2024
|
|
8Q3S
 
 | | HsNMT1 in complex with both MyrCoA and GNCFSKAR inhibitor peptide | | Descriptor: | CHLORIDE ION, COENZYME A, GLY-ASN-CYS-PHE-SER-LYS-ALA-ARG, ... | | Authors: | Dian, C, Giglione, C, Meinnel, T. | | Deposit date: | 2023-08-04 | | Release date: | 2024-08-14 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Novel, tightly structurally related N-myristoyltransferase inhibitors display equally potent yet distinct inhibitory mechanisms.
Structure, 32, 2024
|
|
8Q23
 
 | | HsNMT1 in complex with both MyrCoA and Ac-D-ORN-SFSKPR inhibitor peptide | | Descriptor: | ACE-ORD-SER-PHE-SER-LYS-PRO-ARG, CHLORIDE ION, GLYCEROL, ... | | Authors: | Dian, C, Giglione, C, Meinnel, T. | | Deposit date: | 2023-08-01 | | Release date: | 2024-08-14 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Novel, tightly structurally related N-myristoyltransferase inhibitors display equally potent yet distinct inhibitory mechanisms.
Structure, 32, 2024
|
|
8Q3T
 
 | | HsNMT1 in complex with both MyrCoA and GNCFSKPRVPTK inhibitor peptide | | Descriptor: | CHLORIDE ION, COENZYME A, GLYCEROL, ... | | Authors: | Dian, C, Giglione, C, Meinnel, T. | | Deposit date: | 2023-08-04 | | Release date: | 2024-08-14 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Novel, tightly structurally related N-myristoyltransferase inhibitors display equally potent yet distinct inhibitory mechanisms.
Structure, 32, 2024
|
|
