1X9V
 
 | | Dimeric structure of the C-terminal domain of Vpr | | Descriptor: | VPR protein | | Authors: | Bourbigot, S, Beltz, H, Denis, J, Morellet, N, Roques, B.P, Mely, Y, Bouaziz, S. | | Deposit date: | 2004-08-24 | | Release date: | 2005-06-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The C-terminal domain of the HIV-1 regulatory protein Vpr adopts an antiparallel dimeric structure in solution via its leucine-zipper-like domain
Biochem.J., 387, 2005
|
|
1X9W
 
 | | T7 DNA polymerase in complex with a primer/template DNA containing a disordered N-2 aminofluorene on the template, crystallized with dideoxy-ATP as the incoming nucleotide. | | Descriptor: | 5'-D(*CP*CP*CP*(AFG)*AP*TP*CP*AP*CP*AP*CP*TP*AP*CP*CP*AP*AP*TP*CP*AP*CP*TP*CP*TP*CP*C)-3', 5'-D(*GP*GP*AP*GP*AP*GP*TP*GP*AP*TP*T*GP*GP*T*AP*GP*TP*GP*TP*GP*AP*(2DT))-3', DNA polymerase, ... | | Authors: | Dutta, S, Li, Y, Johnson, D, Dzantiev, L, Richardson, C.C, Romano, L.J, Ellenberger, T. | | Deposit date: | 2004-08-24 | | Release date: | 2004-10-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of 2-acetylaminofluorene and 2-aminofluorene in complex with T7 DNA polymerase reveal mechanisms of mutagenesis.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1X9X
 
 | | Solution Structure of Dimeric SAM Domain from MAPKKK Ste11 | | Descriptor: | Serine/threonine-protein kinase STE11 | | Authors: | Bhattacharjya, S, Xu, P, Gingras, R, Shaykhutdinov, R, Wu, C, Whiteway, M, Ni, F. | | Deposit date: | 2004-08-24 | | Release date: | 2005-08-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the dimeric SAM domain of MAPKKK Ste11 and its interactions with the adaptor protein Ste50 from the budding yeast: implications for Ste11 activation and signal transmission through the Ste50-Ste11 complex.
J.Mol.Biol., 344, 2004
|
|
1X9Y
 
 | | The prostaphopain B structure | | Descriptor: | cysteine proteinase | | Authors: | Filipek, R, Szczepanowski, R, Sabat, A, Potempa, J, Bochtler, M. | | Deposit date: | 2004-08-24 | | Release date: | 2004-11-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Prostaphopain B structure: a comparison of proregion-mediated and staphostatin-mediated protease inhibition.
Biochemistry, 43, 2004
|
|
1X9Z
 
 | | Crystal structure of the MutL C-terminal domain | | Descriptor: | CHLORIDE ION, DNA mismatch repair protein mutL, GLYCEROL, ... | | Authors: | Guarne, A, Ramon-Maiques, S, Wolff, E.M, Ghirlando, R, Hu, X, Miller, J.H, Yang, W. | | Deposit date: | 2004-08-24 | | Release date: | 2004-10-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the MutL C-terminal domain: a model of intact MutL and its roles in mismatch repair
Embo J., 23, 2004
|
|
1XA0
 
 | | Crystal Structure of MCSG Target APC35536 from Bacillus stearothermophilus | | Descriptor: | CHLORIDE ION, Putative NADPH Dependent oxidoreductases, SULFATE ION | | Authors: | Brunzelle, J.S, Sommerhalter, M, Minasov, G, Shuvalova, L, Collart, F.R, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-08-24 | | Release date: | 2004-10-05 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of MCSG Target APC35536 from Bacillus stearothermophilus
To be Published
|
|
1XA1
 
 | | Crystal structure of the sensor domain of BlaR1 from Staphylococcus aureus in its apo form | | Descriptor: | PHOSPHATE ION, PYROPHOSPHATE 2-, Regulatory protein blaR1 | | Authors: | Wilke, M.S, Hills, T.L, Zhang, H.Z, Chambers, H.F, Strynadka, N.C. | | Deposit date: | 2004-08-24 | | Release date: | 2004-09-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of the Apo and penicillin-acylated forms of the BlaR1 beta-lactam sensor of Staphylococcus aureus.
J.Biol.Chem., 279, 2004
|
|
1XA2
 
 | |
1XA3
 
 | | Crystal structure of CaiB, a type III CoA transferase in carnitine metabolism | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Crotonobetainyl-CoA:carnitine CoA-transferase, SULFATE ION | | Authors: | Stenmark, P, Gurmu, D, Nordlund, P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-08-25 | | Release date: | 2004-11-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of CaiB, a Type-III CoA Transferase in Carnitine Metabolism
Biochemistry, 43, 2004
|
|
1XA4
 
 | | Crystal structure of CaiB, a type III CoA transferase in carnitine metabolism | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, COENZYME A, Crotonobetainyl-CoA:carnitine CoA-transferase, ... | | Authors: | Stenmark, P, Gurmu, D, Nordlund, P. | | Deposit date: | 2004-08-25 | | Release date: | 2004-11-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of CaiB, a Type-III CoA Transferase in Carnitine Metabolism
Biochemistry, 43, 2004
|
|
1XA5
 
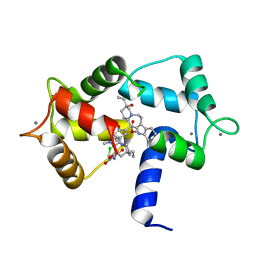 | | Structure of Calmodulin in complex with KAR-2, a bis-indol alkaloid | | Descriptor: | 3"-(BETA-CHLOROETHYL)-2",4"-DIOXO-3, 5"-SPIRO-OXAZOLIDINO-4-DEACETOXY-VINBLASTINE, CALCIUM ION, ... | | Authors: | Horvath, I, Harmat, V, Hlavanda, E, Naray-Szabo, G, Ovadi, J. | | Deposit date: | 2004-08-25 | | Release date: | 2004-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | The structure of the complex of calmodulin with KAR-2: a novel mode of binding explains the unique pharmacology of the drug
J.Biol.Chem., 280, 2005
|
|
1XA6
 
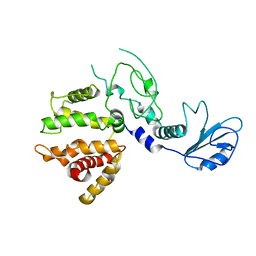 | | Crystal Structure of the Human Beta2-Chimaerin | | Descriptor: | Beta2-chimaerin, ZINC ION | | Authors: | Canagarajah, B, Leskow, F.C, Ho, J.Y, Mischak, H, Saidi, L.F, Kazanietz, M.G, Hurley, J.H. | | Deposit date: | 2004-08-25 | | Release date: | 2004-11-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural mechanism for lipid activation of the Rac-specific GAP, beta2-chimaerin.
Cell(Cambridge,Mass.), 119, 2004
|
|
1XA7
 
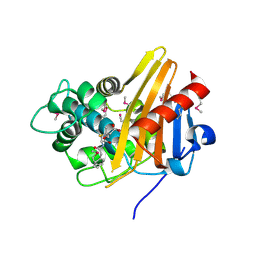 | | Crystal structure of the benzylpenicillin-acylated BlaR1 sensor domain from Staphylococcus aureus | | Descriptor: | OPEN FORM - PENICILLIN G, Regulatory protein BlaR1 | | Authors: | Wilke, M.S, Hills, T.L, Zhang, H.Z, Chambers, H.F, Strynadka, N.C. | | Deposit date: | 2004-08-25 | | Release date: | 2004-09-21 | | Last modified: | 2018-10-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of the Apo and penicillin-acylated forms of the BlaR1 beta-lactam sensor of Staphylococcus aureus.
J.Biol.Chem., 279, 2004
|
|
1XA8
 
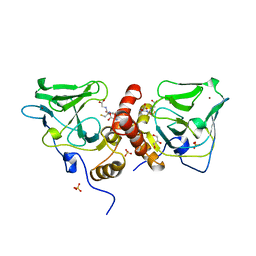 | | Crystal Structure Analysis of Glutathione-dependent formaldehyde-activating enzyme (Gfa) | | Descriptor: | GLUTATHIONE, GLYCEROL, Glutathione-dependent formaldehyde-activating enzyme, ... | | Authors: | Neculai, A.M, Neculai, D, Griesinger, C, Vorholt, J.A, Becker, S. | | Deposit date: | 2004-08-25 | | Release date: | 2004-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A dynamic zinc redox switch
J.Biol.Chem., 280, 2005
|
|
1XA9
 
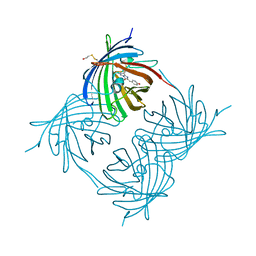 | | Crystal structure of yellow fluorescent protein zFP538 K66M green mutant | | Descriptor: | BETA-MERCAPTOETHANOL, fluorescent protein FP538 | | Authors: | Remington, S.J, Wachter, R.M, Yarbrough, D.K, Branchaud, B, Anderson, D.C, Kallio, K, Lukyanov, K.A. | | Deposit date: | 2004-08-25 | | Release date: | 2005-02-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | zFP538, a yellow-fluorescent protein from Zoanthus, contains a novel three-ring chromophore.
Biochemistry, 44, 2005
|
|
1XAA
 
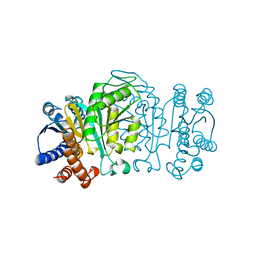 | |
1XAB
 
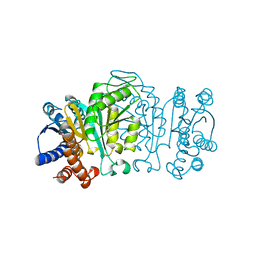 | |
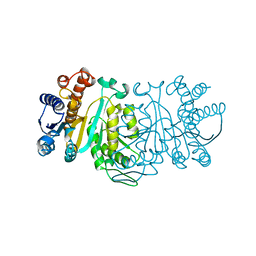
1XAC
 
 | |
1XAD
 
 | |
1XAE
 
 | | Crystal structure of wild type yellow fluorescent protein zFP538 from Zoanthus | | Descriptor: | BETA-MERCAPTOETHANOL, fluorescent protein FP538 | | Authors: | Remington, S.J, Wachter, R.M, Yarbrough, D.K, Branchaud, B, Anderson, D.C, Kallio, K, Lukyanov, K.A. | | Deposit date: | 2004-08-25 | | Release date: | 2005-02-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | zFP538, a yellow-fluorescent protein from Zoanthus, contains a novel three-ring chromophore.
Biochemistry, 44, 2005
|
|
1XAF
 
 | | Crystal Structure of Protein of Unknown Function YfiH from Shigella flexneri 2a str. 2457T | | Descriptor: | ACETATE ION, GLYCEROL, ZINC ION, ... | | Authors: | Kim, Y, Maltseva, N, Dementieva, I, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-08-25 | | Release date: | 2004-08-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of hypothetical protein YfiH from Shigella flexneri at 2 A resolution.
Proteins, 63, 2006
|
|
1XAG
 
 | | CRYSTAL STRUCTURE OF STAPHLYOCOCCUS AUREUS 3-DEHYDROQUINATE SYNTHASE (DHQS) IN COMPLEX WITH ZN2+, NAD+ AND CARBAPHOSPHONATE | | Descriptor: | 3-dehydroquinate synthase, CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Nichols, C.E, Ren, J, Leslie, K, Dhaliwal, B, Lockyer, M, Charles, I, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2004-08-25 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Comparison of ligand induced conformational changes and domain closure mechanisms, between prokaryotic and eukaryotic dehydroquinate synthases.
J.Mol.Biol., 343, 2004
|
|
1XAH
 
 | | CRYSTAL STRUCTURE OF STAPHLYOCOCCUS AUREUS 3-DEHYDROQUINATE SYNTHASE (DHQS) IN COMPLEX WITH ZN2+ AND NAD+ | | Descriptor: | 3-dehydroquinate synthase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | Authors: | Nichols, C.E, Ren, J, Leslie, K, Dhaliwal, B, Lockyer, M, Charles, I, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2004-08-25 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Comparison of ligand induced conformational changes and domain closure mechanisms, between prokaryotic and eukaryotic dehydroquinate synthases.
J.Mol.Biol., 343, 2004
|
|
1XAI
 
 | | CRYSTAL STRUCTURE OF STAPHLYOCOCCUS AUREUS 3-DEHYDROQUINATE SYNTHASE (DHQS) IN COMPLEX WITH ZN2+, NAD+ AND CARBAPHOSPHONATE | | Descriptor: | 3-dehydroquinate synthase, ZINC ION, [1R-(1ALPHA,3BETA,4ALPHA,5BETA)]-5-(PHOSPHONOMETHYL)-1,3,4-TRIHYDROXYCYCLOHEXANE-1-CARBOXYLIC ACID | | Authors: | Nichols, C.E, Ren, J, Leslie, K, Dhaliwal, B, Lockyer, M, Charles, I, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2004-08-25 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Comparison of ligand induced conformational changes and domain closure mechanisms, between prokaryotic and eukaryotic dehydroquinate synthases.
J.Mol.Biol., 343, 2004
|
|
1XAJ
 
 | | CRYSTAL STRUCTURE OF STAPHLYOCOCCUS AUREUS 3-DEHYDROQUINATE SYNTHASE (DHQS) IN COMPLEX WITH ZN2+, NAD+ AND CARBAPHOSPHONATE | | Descriptor: | 3-dehydroquinate synthase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION, ... | | Authors: | Nichols, C.E, Ren, J, Leslie, K, Dhaliwal, B, Lockyer, M, Charles, I, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2004-08-25 | | Release date: | 2005-03-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Comparison of ligand induced conformational changes and domain closure mechanisms, between prokaryotic and eukaryotic dehydroquinate synthases.
J.Mol.Biol., 343, 2004
|
|
